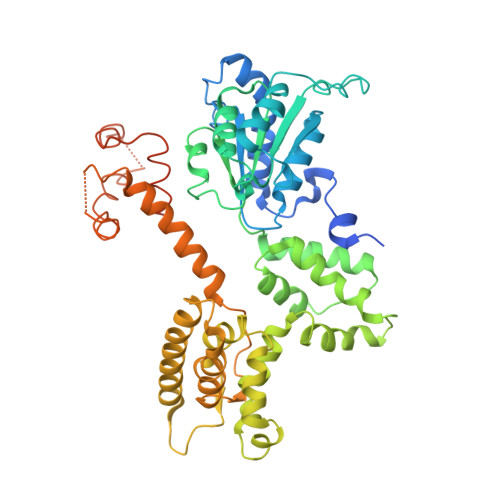
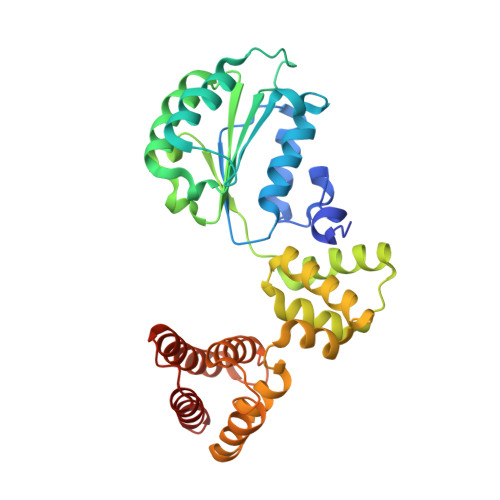
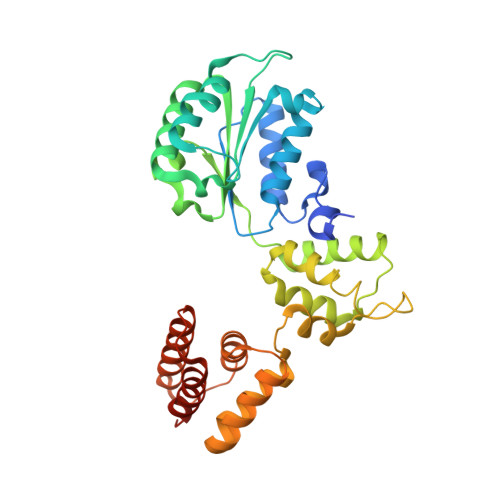
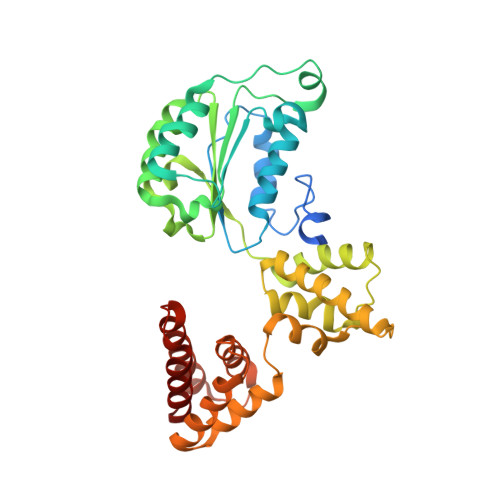
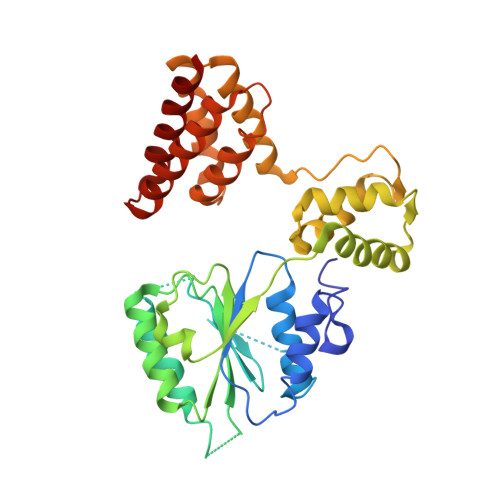
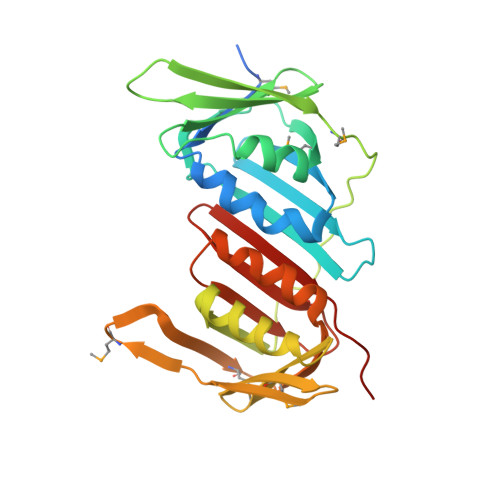
Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Bowman, G.D., O'Donnell, M., Kuriyan, J.(2004) Nature 429: 724-730
- PubMed: 15201901
- DOI: https://doi.org/10.1038/nature02585
- Primary Citation of Related Structures:
1SXJ - PubMed Abstract:
Sliding clamps are ring-shaped proteins that encircle DNA and confer high processivity on DNA polymerases. Here we report the crystal structure of the five-protein clamp loader complex (replication factor-C, RFC) of the yeast Saccharomyces cerevisiae, bound to the sliding clamp (proliferating cell nuclear antigen, PCNA). Tight interfacial coordination of the ATP analogue ATP-gammaS by RFC results in a spiral arrangement of the ATPase domains of the clamp loader above the PCNA ring. Placement of a model for primed DNA within the central hole of PCNA reveals a striking correspondence between the RFC spiral and the grooves of the DNA double helix. This model, in which the clamp loader complex locks onto primed DNA in a screw-cap-like arrangement, provides a simple explanation for the process by which the engagement of primer-template junctions by the RFC:PCNA complex results in ATP hydrolysis and release of the sliding clamp on DNA.
- Howard Hughes Medical Institute, Department of Molecular and Cell Biology and Department of Chemistry, University of California, Berkeley, California 94720, USA.
Organizational Affiliation: