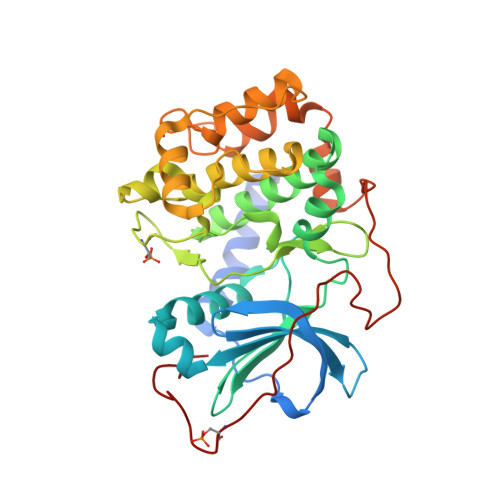
Structure-based optimization of novel azepane derivatives as PKB inhibitors
Breitenlechner, C.B., Wegge, T., Berillon, L., Graul, K., Marzenell, K., Friebe, W.G., Thomas, U., Schumacher, R., Huber, R., Engh, R.A., Masjost, B.(2004) J Med Chem 47: 1375-1390
- PubMed: 14998327
- DOI: https://doi.org/10.1021/jm0310479
- Primary Citation of Related Structures:
1SVE, 1SVG, 1SVH, 1VEB - PubMed Abstract:
Novel azepane derivatives were prepared and evaluated for protein kinase B (PKB-alpha) and protein kinase A (PKA) inhibition. The original (-)-balanol-derived lead structure (4R)-4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-benzoic acid (3R)-3-[(pyridine-4-carbonyl)amino]-azepan-4-yl ester (1) (IC(50) (PKB-alpha) = 5 nM) which contains an ester moiety was found to be plasma unstable and therefore unsuitable as a drug. Based upon molecular modeling studies using the crystal structure of the complex between PKA and 1, the five compounds N-[(3R,4R)-4-[4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-benzoylamino]-azepan-3-yl]-isonicotinamide (4), (3R,4R)-N-[4-[4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-benzyloxy]-azepan-3-yl]-isonicotinamide (5), N-[(3R,4S)-4-[4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-phenylamino]-methyl]-azepan-3-yl)-isonicotinamide (6), N-[(3R,4R)-4-[4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-benzylamino]-azepan-3-yl]-isonicotinamide (7), and N-[(3R,4S)-4-(4-[trans-2-[4-(2-fluoro-6-hydroxy-3-methoxy-benzoyl)-phenyl]-vinyl]-azepan-3-yl)-isonicotinamide (8) with linkers isosteric to the ester were designed, synthesized, and tested for in vitro inhibitory activity against PKA and PKB-alpha and for plasma stability in mouse plasma.(1) Compound 4 was found to be plasma stable and highly active (IC(50) (PKB-alpha) = 4 nM). Cocrystals with PKA were obtained for 4, 5, and 8 and analyzed for binding interactions and conformational changes in the ligands and protein in order to rationalize the different activities of the molecules.
- Pharma Research, Roche Diagnostics GmbH, Werk Penzberg, Nonnenwald 2, D-82372 Penzberg, Germany. birgit.masjost@roche.com
Organizational Affiliation: