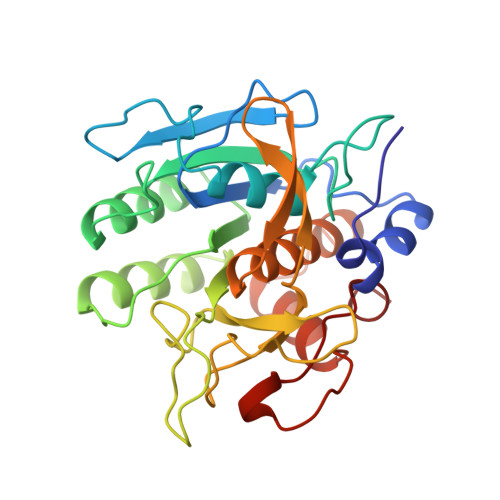
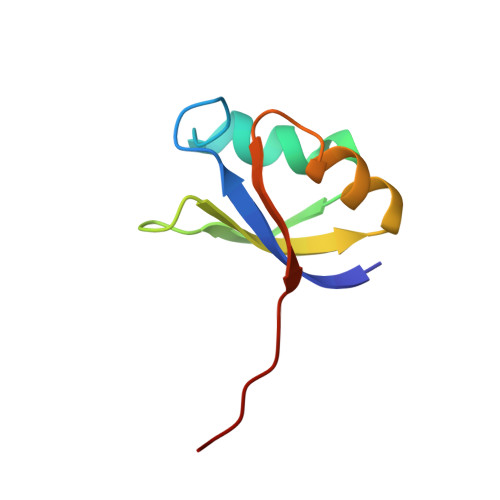
The crystal structure of an autoprocessed Ser221Cys-subtilisin E-propeptide complex at 2.0 A resolution.
Jain, S.C., Shinde, U., Li, Y., Inouye, M., Berman, H.M.(1998) J Mol Biology 284: 137-144
- PubMed: 9811547
- DOI: https://doi.org/10.1006/jmbi.1998.2161
- Primary Citation of Related Structures:
1SCJ - PubMed Abstract:
We report here the crystallographic structure determination of an autoprocessed (Ser221Cys)-subtilisin E-propeptide complex at 2.0 A resolution. The subtilisin domain sequence has a single substitution (Ser221Cys) which has been shown to block the maturation process prior to degradation of the propeptide domain (77 residues) that acts as an intramolecular chaperon. This mutation, however, did not prevent the enzyme from cleaving its propeptide domain with a 60-80% efficiency. The current determination is the first example of a subtilisin E-propeptide complex which has been autoprocessed. A previous structure determination of a BPN'-prosegment complex has been reported in which the subtilisin domain was extensively mutated and a calcium binding loop was deleted. Further, in this earlier determination, the complex was formed by the addition of separately expressed propeptide domain. The structure determination reported here provides additional information about the nature of the interaction between the subtilisin and propeptide domains in this complex.
- Department of Chemistry, Rutgers University, 610 Taylor Road, Piscataway, NJ, 08854-8087, USA.
Organizational Affiliation: