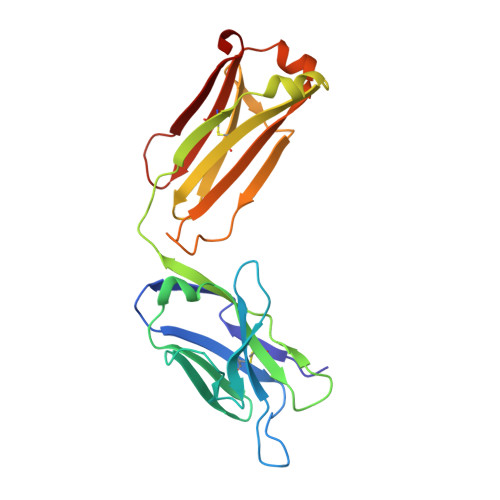
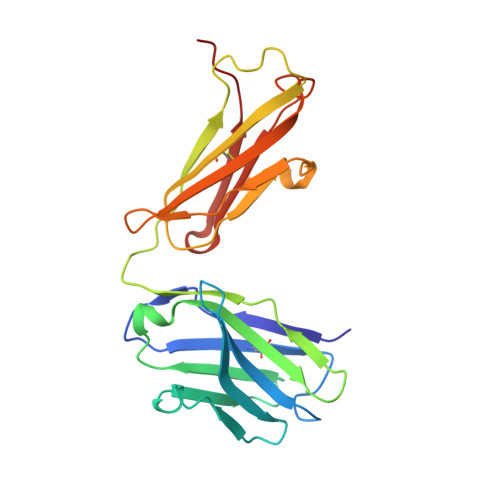
[A new crystal form of the Fab fragment of a monoclonal antibody to human interleukin-2: the three-dimensional structure at 2.7 A resolution].
Pletnev, V.Z., Goriacheva, E.A., Tsygannik, I.N., Nesmeianov, V.A., Pletnev, S.V., Pangborn, W., Daux, W.(null) Bioorg Khim 30: 466-469
- PubMed: 15562966
- DOI: https://doi.org/10.1023/b:rubi.0000043783.73562.ef
- Primary Citation of Related Structures:
1S5I - PubMed Abstract:
The three-dimensional structure of the antigen-binding fragment of a monoclonal antibody to human interleukin-2 in a new crystal form (space group P2(1)2(1)2(1); unit cell parameters: a = 42.82, b = 90.68, and c = 139.82 A) was determined by the X-ray molecular replacement method at the resolution of 2.7 A. The protein folding and the stereochemistry of its antigen-binding site were comparatively analyzed. The English version of the paper: Russian Journal of Bioorganic Chemistry, 2004, vol. 30, no. 5; see also http: // www.maik.ru.