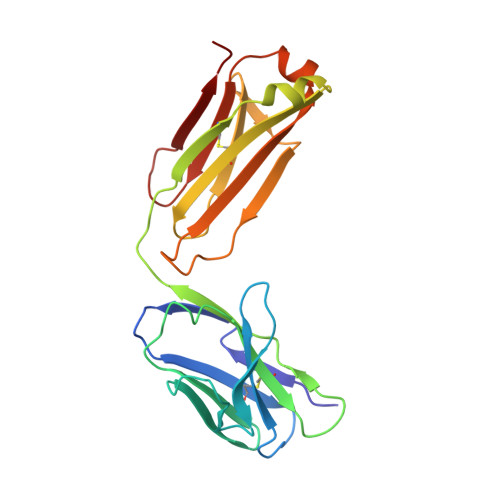
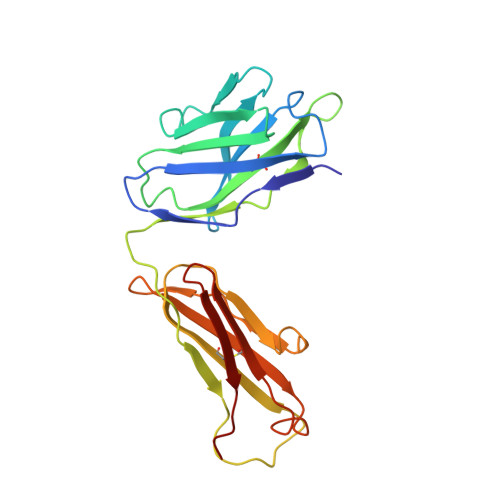
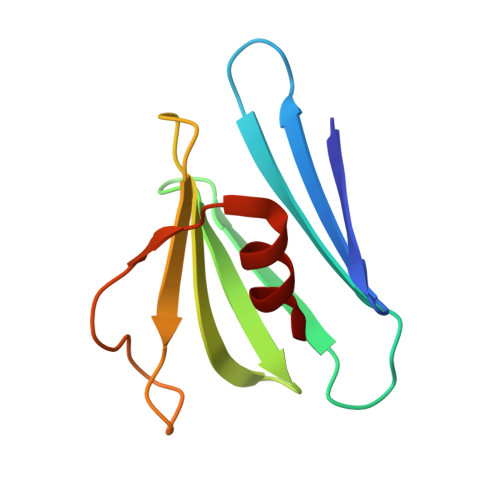
Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
Becker, M., Bunikis, J., Lade, B.D., Dunn, J.J., Barbour, A.G., Lawson, C.L.(2005) J Biological Chem 280: 17363-17370
- PubMed: 15713683
- DOI: https://doi.org/10.1074/jbc.M412842200
- Primary Citation of Related Structures:
1P4P, 1RJL - PubMed Abstract:
Certain antibody Fab fragments directed against the C terminus of outer surface protein B (OspB), a major lipoprotein of the Lyme disease spirochete, Borrelia burgdorferi, have the unusual property of being bactericidal even in the absence of complement. We report here x-ray crystal structures of a C-terminal fragment of B. burgdorferi OspB, which spans residues 152-296, alone at 2.0-A resolution, and in a complex with the bactericidal Fab H6831 at 2.6-A resolution. The H6831 epitope is topologically analogous to the LA-2 epitope of OspA and is centered around OspB Lys-253, a residue essential for H6831 recognition. A beta-sheet present in the free OspB fragment is either disordered or removed by proteolysis in the H6831-bound complex. Other conformational changes between free and H6831-bound structures are minor and appear to be related to this loss. In both crystal structures, OspB C-terminal fragments form artificial dimers connected by intermolecular beta-sheets. OspB structure, stability, and possible mechanisms of killing by H6831 and other bactericidal Fabs are discussed in light of the structural data.
- Biology Department, Brookhaven National Laboratory, Upton, New York 11973, USA. mbecker@bnl.gov
Organizational Affiliation: