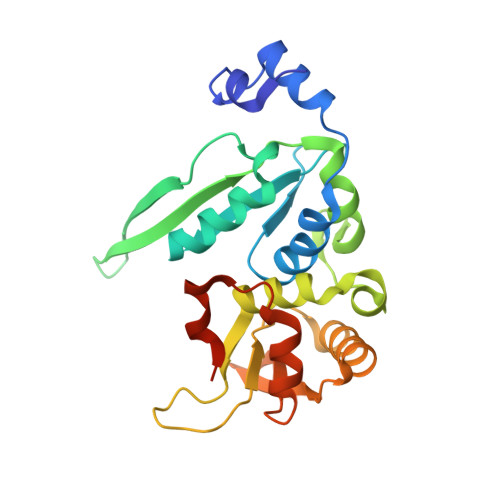
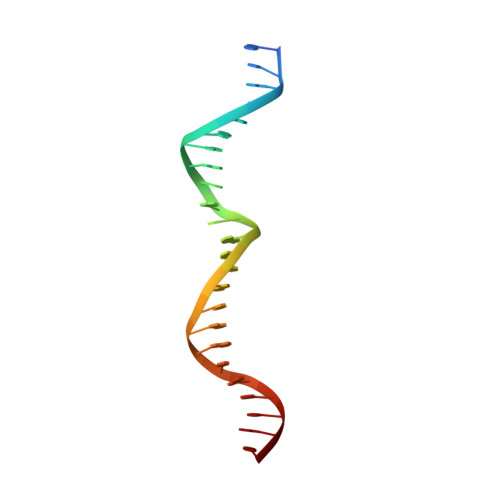
The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity
Moure, C.M., Gimble, F.S., Quiocho, F.A.(2003) J Mol Biology 334: 685-695
- PubMed: 14636596
- DOI: https://doi.org/10.1016/j.jmb.2003.09.068
- Primary Citation of Related Structures:
1R7M - PubMed Abstract:
The I-SceI homing endonuclease enhances gene targeting by introducing double-strand breaks at specific chromosomal loci, thereby increasing the recombination frequency. Here, we report the crystal structure of the enzyme complexed to its DNA substrate and Ca(2+) determined at 2.25A resolution. The structure shows the prototypical beta-saddle of LAGLIDADG homing endonucleases that is contributed by two pseudo-symmetric domains. The high specificity of I-SceI is explained by the large number of protein-DNA contacts, many that are made by a long beta-hairpin loop that reaches into the major groove of the DNA. The DNA minor groove is compressed at the catalytic center, bringing the two scissile phosphodiester bonds into close proximity. The protein-Ca(2+)-DNA structure shows the protein bound to its DNA substrate in a pre-reactive state that is defined by the presence of two asymmetric active sites, one of which appears poised to first cleave the DNA bottom strand.
- Howard Hughes Medical Institute, Baylor College of Medicine, Houston, TX 77030, USA.
Organizational Affiliation: