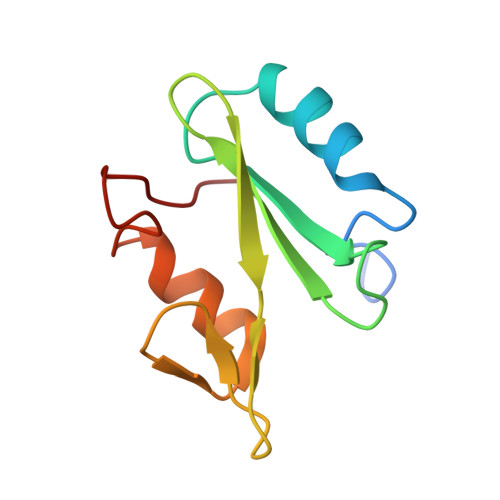
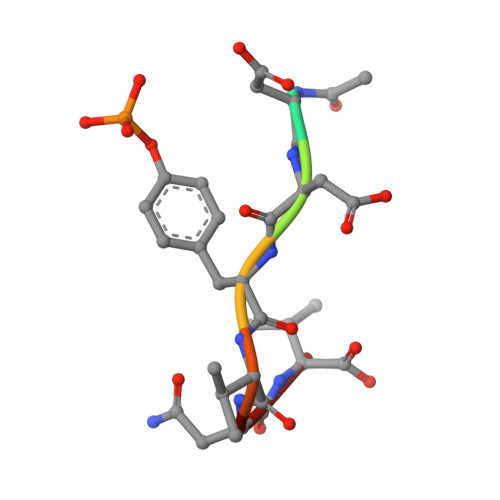
Structural basis for differential recognition of tyrosine-phosphorylated sites in the linker for activation of T cells (LAT) by the adaptor Gads.
Cho, S., Velikovsky, C.A., Swaminathan, C.P., Houtman, J.C., Samelson, L.E., Mariuzza, R.A.(2004) EMBO J 23: 1441-1451
- PubMed: 15029250
- DOI: https://doi.org/10.1038/sj.emboj.7600168
- Primary Citation of Related Structures:
1R1P, 1R1Q, 1R1S - PubMed Abstract:
The transmembrane protein, linker for activation of T cells (LAT), is essential for T-cell activation and development. Phosphorylation of LAT at multiple tyrosines creates binding sites for the adaptors Gads and Grb2, leading to nucleation of multiprotein signaling complexes. Human LAT contains five potential binding sites for Gads, of which only those at Tyr171 and Tyr191 appear necessary for T-cell function. We asked whether Gads binds preferentially to these sites, as differential recognition could assist in assembling defined LAT-based complexes. Measured calorimetrically, Gads-SH2 binds LAT tyrosine phosphorylation sites 171 and 191 with higher affinities than the other sites, with the differences ranging from only several fold weaker binding to no detectable interaction. Crystal structures of Gads-SH2 complexed with phosphopeptides representing sites 171, 191 and 226 were determined to 1.8-1.9 A resolutions. The structures reveal the basis for preferential recognition of specific LAT sites by Gads, as well as for the relatively greater promiscuity of the related adaptor Grb2, whose binding also requires asparagine at position +2 C-terminal to the phosphorylated tyrosine.
- Center for Advanced Research in Biotechnology, WM Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Rockville, MD, USA.
Organizational Affiliation: