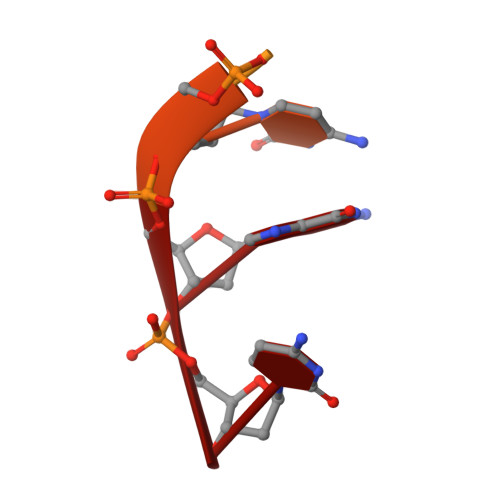
Structural elucidation of the binding and inhibitory properties of lanthanide (III) ions at the 3'-5' exonucleolytic active site of the Klenow fragment
Brautigam, C.A., Aschheim, K., Steitz, T.A.(1999) Chem Biol 6: 901-908
- PubMed: 10631518
- DOI: https://doi.org/10.1016/s1074-5521(00)80009-5
- Primary Citation of Related Structures:
1QSL - PubMed Abstract:
Biochemical and biophysical experiments have shown that two catalytically essential divalent metal ions (termed 'A' and 'B') bind to the 3'-5' exonuclease active site of the Klenow fragment (KF) of Escherichia coli DNA polymerase I. X-ray crystallographic studies have established the normal positions in the KF 3'-5' exonuclease (KF exo) active site of the two cations and the single-stranded DNA substrate. Lanthanide (III) luminescence studies have demonstrated, however, that only a single europium (III) ion (Eu3+) binds to the KF exo active site. Furthermore, Eu3+ does not support catalysis by KF exo or several other two-metal-ion phosphoryl-transfer enzymes. A crystal structure of KF complexed with both Eu3+ and substrate single-stranded oligodeoxynucleotide shows that a lone Eu3+ is bound near to metal-ion site A. Comparison of this structure to a relevant native structure reveals that the bound Eu3+ causes a number of changes to the KF exo active site. The scissile phosphate of the substrate is displaced from its normal position by about 1 A when Eu3+ is bound and the presence of Eu3+ in the active site precludes the binding of the essential metal ion B. The substantial, lanthanide-induced differences in metal-ion and substrate binding to KF exo account for the inhibition of this enzyme by Eu3+. These changes also explain the inability of KF exo to bind more than one cation in the presence of lanthanides. The mechanistic similarity between KF exo and other two-metal-ion phosphoryl-transfer enzymes suggests that the principles of lanthanide (III) ion binding and inhibition ascertained from this study will probably apply to most members of this class of enzymes.
- Department of Molecular Biophysics and Biochemistry, Howard Hughes Medical Institute, Yale University, New Haven, CT 06520-8114, USA.
Organizational Affiliation: