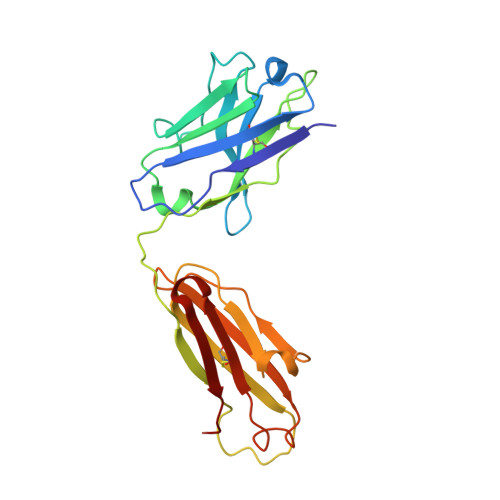
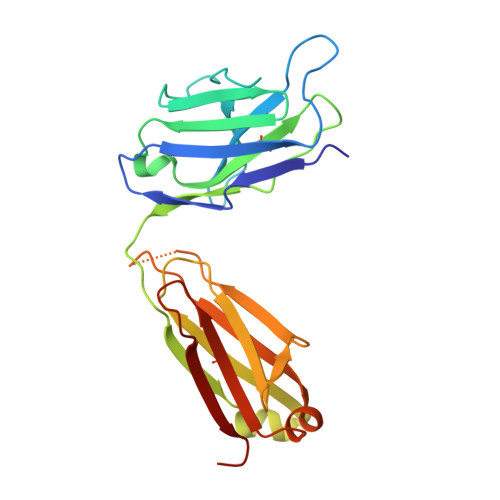
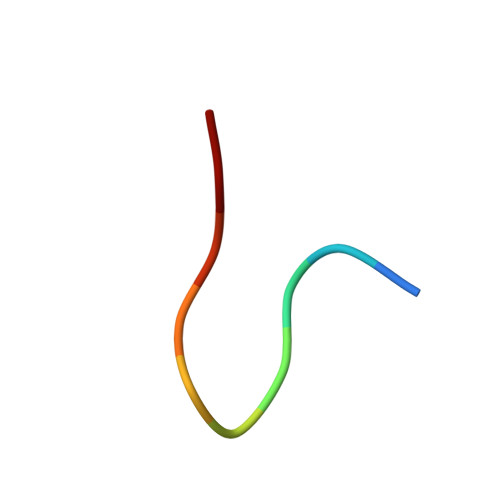Crystal Structure of an Fab Fragment in Complex with a Meningococcal Serosubtype Antigen and a Protein G Domain
Derrick, J.P., Feavers, I.M., Maiden, M.C.J.(1999) J Mol Biology 293: 81
- PubMed: 10512717
- DOI: https://doi.org/10.1006/jmbi.1999.3144
- Primary Citation of Related Structures:
1QKZ - PubMed Abstract:
Many pathogens present highly variable surface proteins to their host as a means of evading immune responses. The structure of a peptide antigen corresponding to the subtype P1.7 variant of the porin PorA from the human pathogen Neisseria meningitidis was determined by solution of the X-ray crystal structure of the ternary complex of the peptide (ANGGASGQVK) in complex with a Fab fragment and a domain from streptococcal protein G to 1.95 A resolution. The peptide adopted a beta-hairpin structure with a type I beta-turn between residues Gly4P and Gly7P, the conformation of the peptide being further stabilised by a pair of hydrogen bonds from the side-chain of Asn2P to main-chain atoms in Val9P. The antigen binding site within the Fab formed a distinct crevice lined by a high proportion of apolar amino acids. Recognition was supplemented by hydrogen bonds from heavy chain residues Thr50H, Asp95H, Leu97H and Tyr100H to main-chain and side-chain atoms in the peptide. Complementarity-determining region (CDR) 3 of the heavy chain was responsible for approximately 50 % of the buried surface area formed by peptide-Fab binding, with the remainder made up from CDRs 1 and 3 of the light chain and CDRs 1 and 2 of the heavy chain. Knowledge of the structures of variable surface antigens such as PorA is an essential prerequisite to a molecular understanding of antigenic variation and its implications for vaccine design.
- Department of Biomolecular Sciences, UMIST, Manchester, M60 1QD, UK. Jeremy.Derrick@umist.ac.uk
Organizational Affiliation: