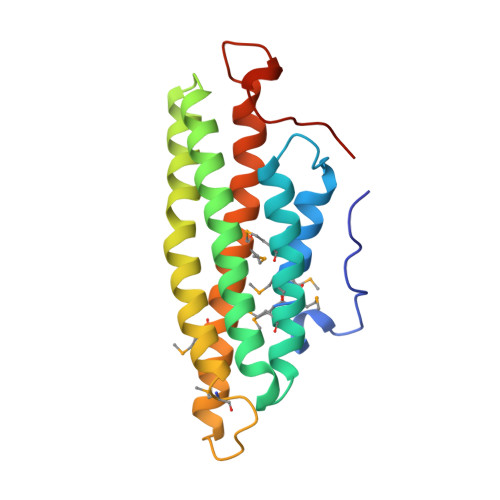
Crystal Structure of the Vinculin Tail and a Pathway for Activation
Bakolitsa, C., De Pereda, J.M., Bagshaw, C.R., Critchley, D.R., Liddington, R.C.(1999) Cell 99: 603
- PubMed: 10612396
- DOI: https://doi.org/10.1016/s0092-8674(00)81549-4
- Primary Citation of Related Structures:
1QKR - PubMed Abstract:
Vinculin plays a dynamic role in the assembly of the actin cytoskeleton. A strong interaction between its head and tail domains that regulates binding to other cytoskeletal components is disrupted by acidic phospholipids. Here, we present the crystal structure of the vinculin tail, residues 879-1066. Five amphipathic helices form an antiparallel bundle that resembles exchangeable apolipoproteins. A C-terminal arm wraps across the base of the bundle and emerges as a hydrophobic hairpin surrounded by a collar of basic residues, adjacent to the N terminus. We show that the C-terminal arm is required for binding to acidic phospholipids but not to actin, and that binding either ligand induces conformational changes that may represent the first step in activation.
- Department of Biochemistry, University of Leicester, United Kingdom.
Organizational Affiliation: