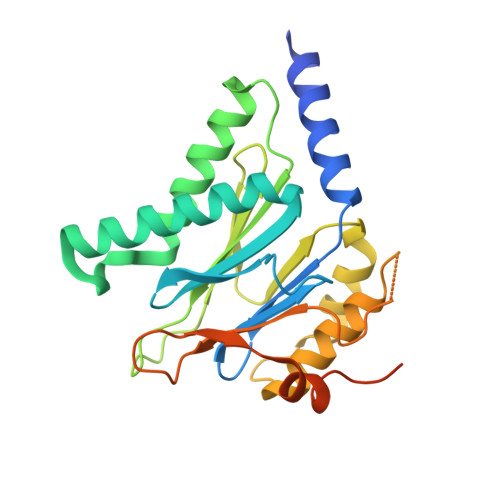
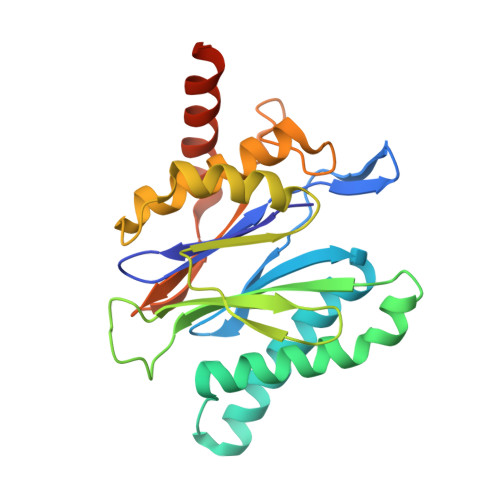
Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
Kwon, Y.D., Nagy, I., Adams, P.D., Baumeister, W., Jap, B.K.(2004) J Mol Biology 335: 233-245
- PubMed: 14659753
- DOI: https://doi.org/10.1016/j.jmb.2003.08.029
- Primary Citation of Related Structures:
1Q5Q, 1Q5R - PubMed Abstract:
To understand the role of the pro-peptide in proteasome assembly, we have determined structures of the Rhodococcus proteasome and a mutant form that prevents the autocatalytic removal of its pro-peptides. The structures reveal that the pro-peptide acts as an assembly-promoting factor by linking its own beta-subunit to two adjacent alpha-subunits, thereby providing a molecular explanation for the observed kinetics of proteasome assembly. The Rhodococcus proteasome has been found to have a substantially smaller contact region between alpha-subunits compared to those regions in the proteasomes of Thermoplasma, yeast, and mammalian cells, suggesting that a smaller contact area between alpha-subunits is likely the structural basis for the Rhodococcus alpha-subunits not assembling into alpha-rings when expressed alone. Analysis of all available beta-subunit structures shows that the contact area between beta-subunits within a beta-ring is not sufficient for beta-ring self-assembly without the additional contact provided by the alpha-ring. This appears to be a fail-safe mechanism ensuring that the active sites on the beta-subunits are activated only after proteasome assembly is complete.
- Graduate Group in Comparative Biochemistry, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: