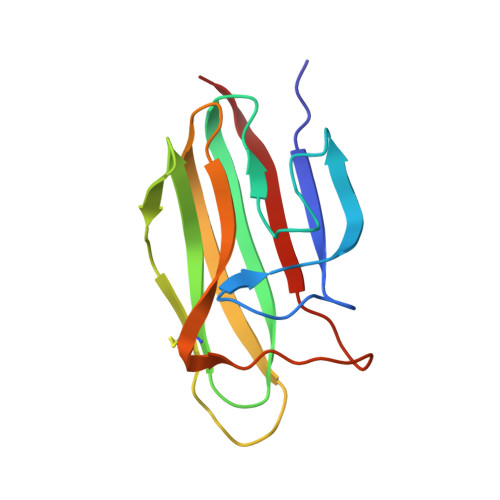
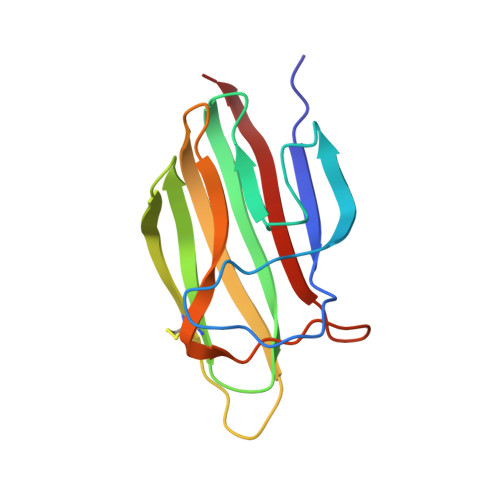
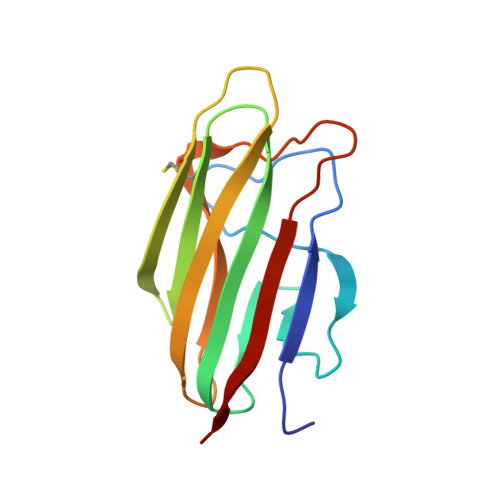
The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties.
Gaboriaud, C., Juanhuix, J., Gruez, A., Lacroix, M., Darnault, C., Pignol, D., Verger, D., Fontecilla-Camps, J.C., Arlaud, G.J.(2003) J Biological Chem 278: 46974-46982
- PubMed: 12960167
- DOI: https://doi.org/10.1074/jbc.M307764200
- Primary Citation of Related Structures:
1PK6 - PubMed Abstract:
C1q is a versatile recognition protein that binds to an amazing variety of immune and non-immune ligands and triggers activation of the classical pathway of complement. The crystal structure of the C1q globular domain responsible for its recognition properties has now been solved and refined to 1.9 A of resolution. The structure reveals a compact, almost spherical heterotrimeric assembly held together mainly by non-polar interactions, with a Ca2+ ion bound at the top. The heterotrimeric assembly of the C1q globular domain appears to be a key factor of the versatile recognition properties of this protein. Plausible three-dimensional models of the C1q globular domain in complex with two of its physiological ligands, C-reactive protein and IgG, are proposed, highlighting two of the possible recognition modes of C1q. The C1q/human IgG1 model suggests a critical role for the hinge region of IgG and for the relative orientation of its Fab domain in C1q binding.
- Laboratoire de Cristallographie et Cristallogéncse des Protéines, Institut de Biologie Structurale Jean-Pierre Ebel, Commissariat à l'Energie Atomique (CEA)-CNRS-Université Joseph Fourier, 41 rue Jules Horowitz, 38027 Grenoble Cedex 1, France.
Organizational Affiliation: