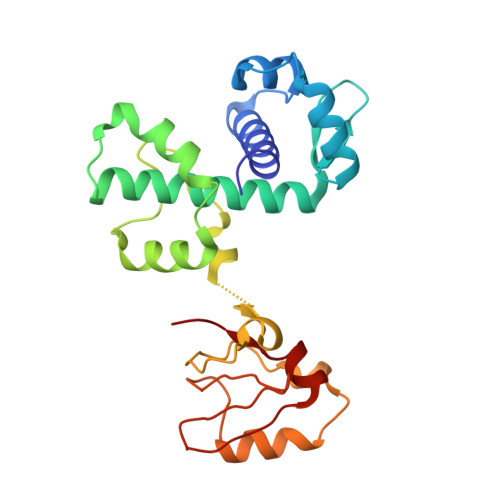
Determinants of the SRC homology domain 3-like fold.
D'Aquino, J.A., Ringe, D.(2003) J Bacteriol 185: 4081-4086
- PubMed: 12837782
- DOI: https://doi.org/10.1128/JB.185.14.4081-4086.2003
- Primary Citation of Related Structures:
1P92 - PubMed Abstract:
In eukaryotes, the Src homology domain 3 (SH3) is a very important motif in signal transduction. SH3 domains recognize poly-proline-rich peptides and are involved in protein-protein interactions. Until now, the existence of SH3 domains has not been demonstrated in prokaryotes. However, the structure of the C-terminal domain of DtxR clearly shows that the fold of this domain is very similar to that of the SH3 domain. In addition, there is evidence that the C-terminal domain of DtxR binds to poly-proline-rich regions. Other bacterial proteins have domains that are structurally similar to the SH3 domain but whose functions are unknown or differ from that of the SH3 domain. The observed similarities between the structures of the C-terminal domain of DtxR and the SH3 domain constitute a perfect system to gain insight into their function and information about their evolution. Our results show that the C-terminal domain of DtxR shares a number of conserved key hydrophobic positions not recognizable from sequence comparison that might be responsible for the integrity of the SH3-like fold. Structural alignment of an ensemble of such domains from unrelated proteins shows a common structural core that seems to be conserved despite the lack of sequence similarity. This core constitutes the minimal requirements of protein architecture for the SH3-like fold.
- Department of Chemistry and Biochemistry and Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, Massachusetts 02454, USA.
Organizational Affiliation: