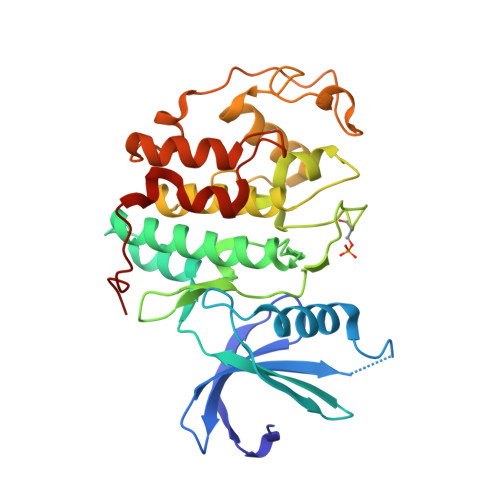
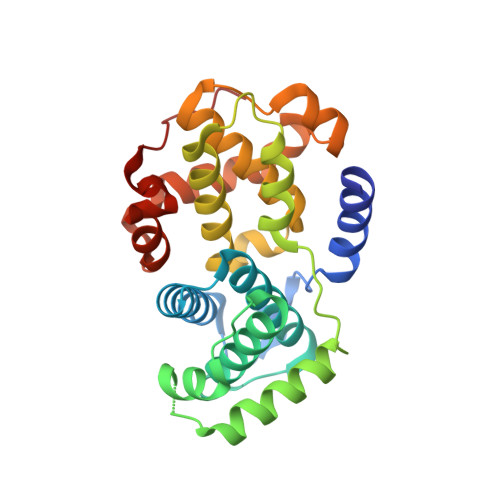
Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Sayle, K.L., Bentley, J., Boyle, F.T., Calvert, A.H., Cheng, Y., Curtin, N.J., Endicott, J.A., Golding, B.T., Hardcastle, I.R., Jewsbury, P., Mesguiche, V., Newell, D.R., Noble, M.E.M., Parsons, R.J., Pratt, D.J., Wang, L.Z., Griffin, R.J.(2003) Bioorg Med Chem Lett 13: 3079
- PubMed: 12941338
- DOI: https://doi.org/10.1016/s0960-894x(03)00651-6
- Primary Citation of Related Structures:
1OGU - PubMed Abstract:
A series of O(4)-cyclohexylmethyl-5-nitroso-6-aminopyrimidines bearing 2-arylamino substituents was synthesised and evaluated for CDK1 and CDK2 inhibitory activity. Consistent with analogous studies with O(6)-cyclohexylmethylpurines, 2-arylaminopyrimidines with a sulfonamide or carboxamide group at the 4'-position were potent inhibitors, with IC(50) values against CDK2 of 1.1+/-0.3 and 34+/-8 nM, respectively. The crystal structure of the 4'-carboxamide derivative, in complex with phospho-Thr160 CDK2/cyclin A, confirmed the expected binding mode of the inhibitor, and revealed an additional interaction between the carboxamide function and an aspartate residue.
- Northern Institute for Cancer Research, School of Natural Sciences-Chemistry, Bedson Building, University of Newcastle, Newcastle Upon Tyne NE1 7RU, UK.
Organizational Affiliation: