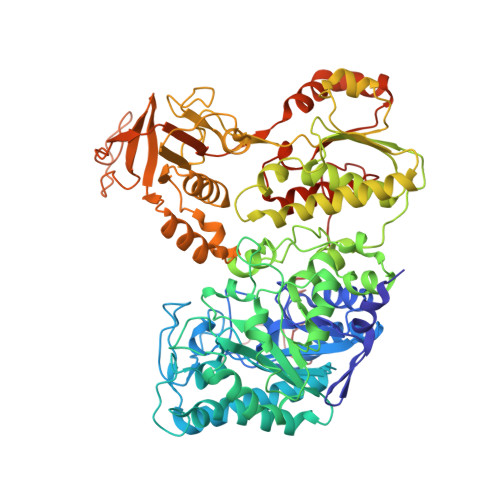
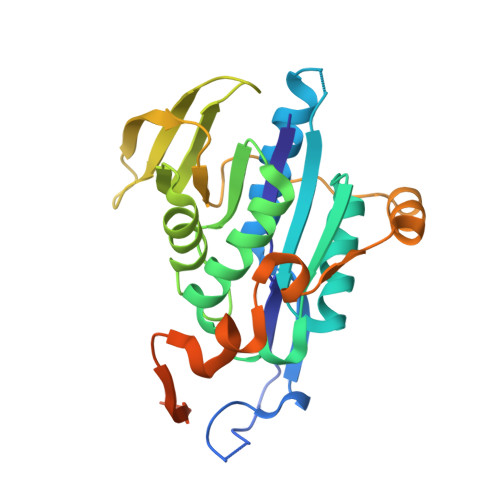
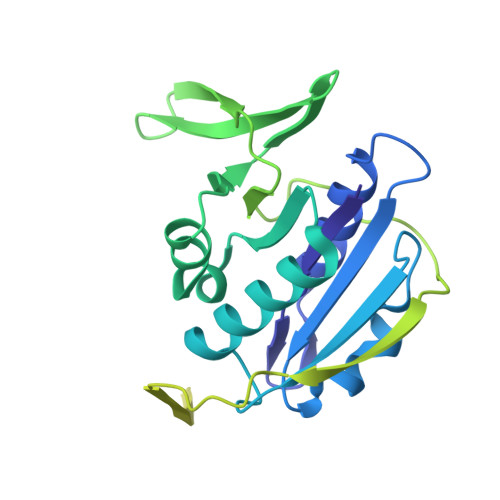
Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Leys, D., Basran, J., Talfournier, F., Sutcliffe, M.J., Scrutton, N.S.(2003) Nat Struct Biol 10: 219
- PubMed: 12567183
- DOI: https://doi.org/10.1038/nsb894
- Primary Citation of Related Structures:
1O94, 1O95, 1O96, 1O97 - PubMed Abstract:
Here we report the crystal structures of a ternary electron transfer complex showing extensive motion at the protein interface. This physiological complex comprises the iron-sulfur flavoprotein trimethylamine dehydrogenase and electron transferring flavoprotein (ETF) from Methylophilus methylotrophus. In addition, we report the crystal structure of free ETF. In the complex, electron density for the FAD domain of ETF is absent, indicating high mobility. Positions for the FAD domain are revealed by molecular dynamics simulation, consistent with crystal structures and kinetic data. A dual interaction of ETF with trimethylamine dehydrogenase provides for dynamical motion at the protein interface: one site acts as an anchor, thereby allowing the other site to sample a large range of interactions, some compatible with rapid electron transfer. This study establishes the role of conformational sampling in multi-domain redox systems, providing insight into electron transfer between ETFs and structurally distinct redox partners.
- Department of Biochemistry, University of Leicester, University Road, Leicester LE1 7RH, UK. dl37@le.ac.uk
Organizational Affiliation: