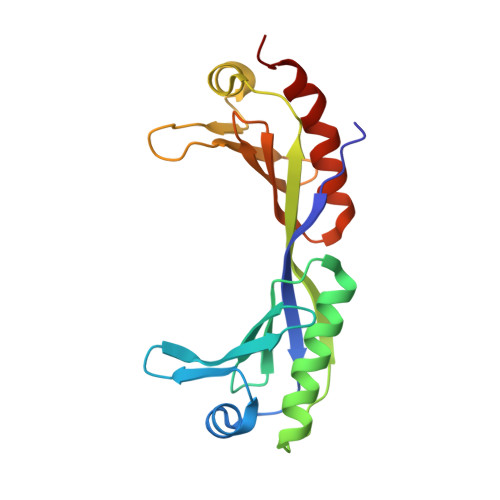
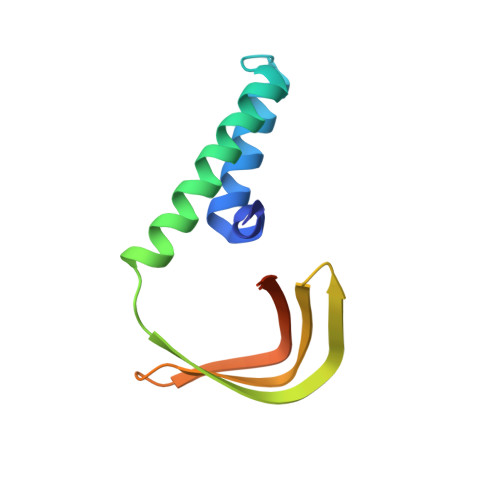
Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes.
Bleichenbacher, M., Tan, S., Richmond, T.J.(2003) J Mol Biology 332: 783-793
- PubMed: 12972251
- DOI: https://doi.org/10.1016/s0022-2836(03)00887-8
- Primary Citation of Related Structures:
1NH2, 1NVP - PubMed Abstract:
RNA polymerase II-dependent transcription requires the assembly of a multi-protein, preinitiation complex on core promoter elements. Transcription factor IID (TFIID) comprising the TATA box-binding protein (TBP) and TBP-associated factors (TAFs) is responsible for promoter recognition in this complex. Subsequent association of TFIIA and TFIIB provides enhanced complex stability. TFIIA is required for transcriptional stimulation by certain viral and cellular activators, and favors formation of the preinitiation complex in the presence of repressor NC2. The X-ray structures of human and yeast TBP/TFIIA/DNA complexes at 2.1A and 1.9A resolution, respectively, are presented here and seen to resemble each other closely. The interactions made by human TFIIA with TBP and DNA within and upstream of the TATA box, including those involving water molecules, are described and compared to the yeast structure. Of particular interest is a previously unobserved region of TFIIA that extends the binding interface with TBP in the yeast, but not in the human complex, and that further elucidates biochemical and genetic results.
- ETH Zürich, Institute for Molecular Biology and Biophysics, ETH-Hönggerberg, CH-8093 Zürich, Switzerland.
Organizational Affiliation: