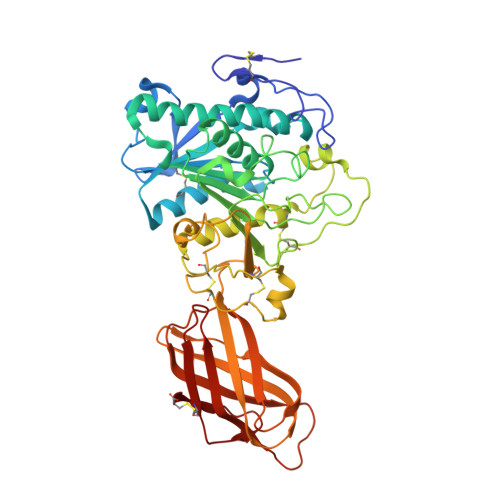
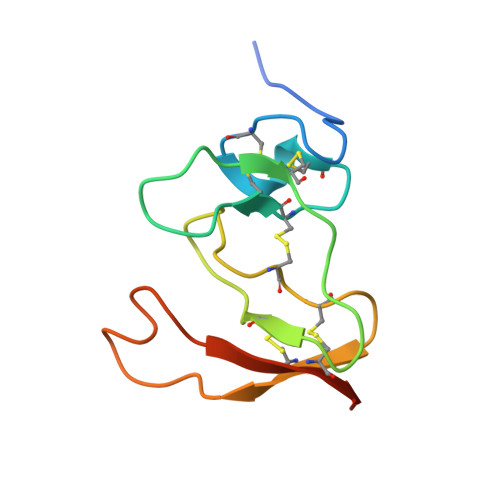
Structure of the pancreatic lipase-procolipase complex
van Tilbeurgh, H., Sarda, L., Verger, R., Cambillau, C.(1992) Nature 359: 159-162
- PubMed: 1522902
- DOI: https://doi.org/10.1038/359159a0
- Primary Citation of Related Structures:
1N8S - PubMed Abstract:
Interfacial adsorption of pancreatic lipase is strongly dependent on the physical chemical properties of the lipid surface. These properties are affected by amphiphiles such as phospholipids and bile salts. In the presence of such amphiphiles, lipase binding to the interface requires a protein cofactor, colipase. We obtained crystals of the pancreatic lipase-procolipase complex and solved the structure at 3.04 A resolution. Here we describe the structure of procolipase, which essentially consists of three 'fingers' and is topologically comparable to snake toxins. The tips of the fingers contain most of the hydrophobic amino acids and presumably form the interfacial binding site. Lipase binding occurs at the opposite side to this site and involves polar interactions. Determination of the three-dimensional structure of pancreatic lipase has revealed the presence of two domains: an amino-terminal domain, at residues 1-336 containing the active site and a carboxy-terminal domain at residues 337-449 (ref. 6). Procolipase binds exclusively to the C-terminal domain of lipase. No conformational change in the lipase molecule is induced by the binding of procolipase.
- LCCMB-CNRS, Faculté de Médecine Nord, Marseille, France.
Organizational Affiliation: