Structure determination of minute virus of mice.
Llamas-Saiz, A.L., Agbandje-McKenna, M., Wikoff, W.R., Bratton, J., Tattersall, P., Rossmann, M.G.(1997) Acta Crystallogr D Biol Crystallogr 53: 93-102
- PubMed: 15299974
- DOI: https://doi.org/10.1107/S0907444996010566
- Primary Citation of Related Structures:
1MVM - PubMed Abstract:
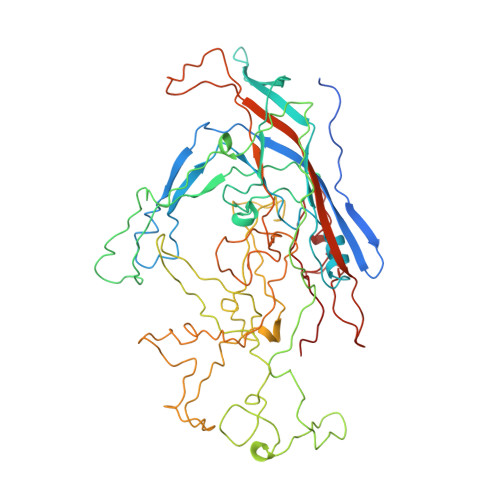
The three-dimensional crystal structure of the single-stranded DNA-containing ('full') parvovirus, minute virus of mice (MVM), has been determined to 3.5 A resolution. Both full and empty particles of MVM were crystallized in the monoclinic space group C2 with cell dimensions of a = 448.7, b = 416.7, c = 305.3 A and beta = 95.8 degrees. Diffraction data were collected at the Cornell High Energy Synchrotron Source using an oscillation camera. The crystals have a pseudo higher R32 space group in which the particles are situated at two special positions with 32 point symmetry, separated by (1/2)c in the hexagonal setting. The self-rotation function showed that the particles are rotated with respect to each other by 60 degrees around the pseudo threefold axis. Subsequently, a more detailed analysis of the structure amplitudes demonstrated that the correct space-group symmetry is C2 as given above. Only one of the three twofold axes perpendicular to the threefold axis in the pseudo R32 space group is a 'true' crystallographic twofold axis; the other two are only 'local' non-crystallographic symmetry axes. The known canine parvovirus (CPV) structure was used as a phasing model to initiate real-space electron-density averaging phase improvement. The electron density was easily interpretable and clearly showed the amino-acid differences between MVM and CPV, although the final overall correlation coefficient was only 0.63. The structure of MVM has a large amount of icosahedrally ordered DNA, amounting to 22% of the viral genome, which is significantly more than that found in CPV.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907-1392, USA.
Organizational Affiliation: