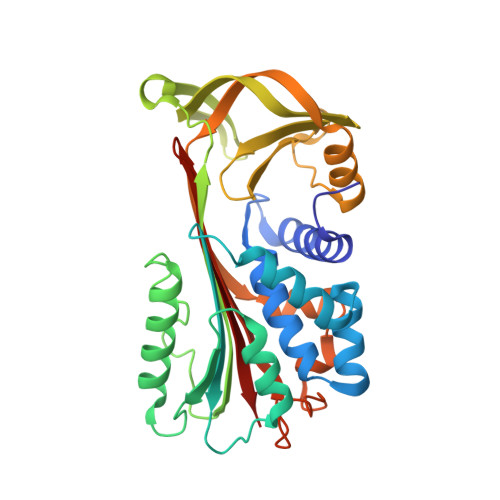
The 1.5 A crystal structure of a prokaryote serpin: controlling conformational change in a heated environment
Irving, J.A., Cabrita, L.D., Rossjohn, J., Pike, R.N., Bottomley, S.P., Whisstock, J.C.(2003) Structure 11: 387-397
- PubMed: 12679017
- DOI: https://doi.org/10.1016/s0969-2126(03)00057-1
- Primary Citation of Related Structures:
1MTP - PubMed Abstract:
Serpins utilize conformational change to inhibit target proteinases; the price paid for this conformational flexibility is that many undergo temperature-induced polymerization. Despite this thermolability, serpins are present in the genomes of thermophilic prokaryotes, and here we characterize the first such serpin, thermopin. Thermopin is a proteinase inhibitor and, in comparison with human alpha(1)-antitrypsin, possesses enhanced stability at 60 degrees C. The 1.5 A crystal structure reveals novel structural features in regions implicated in serpin folding and stability. Thermopin possesses a C-terminal "tail" that interacts with the top of the A beta sheet and plays an important role in the folding/unfolding of the molecule. These data provide evidence as to how this unusual serpin has adapted to fold and function in a heated environment.
- The Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, 3800, Clayton, Australia.
Organizational Affiliation: