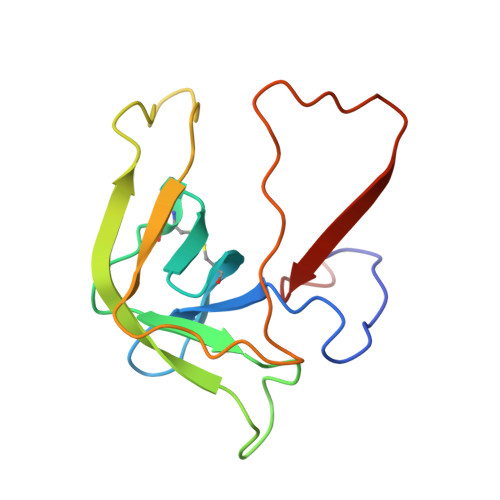
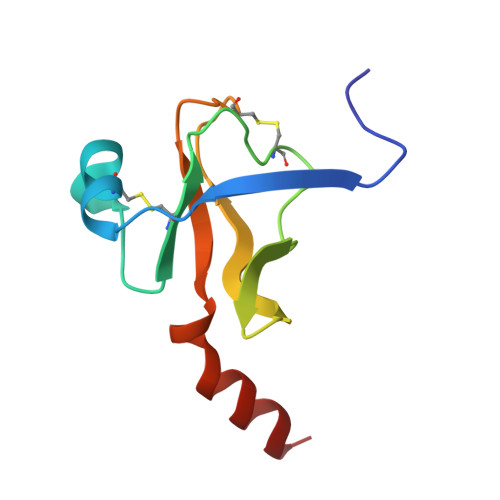
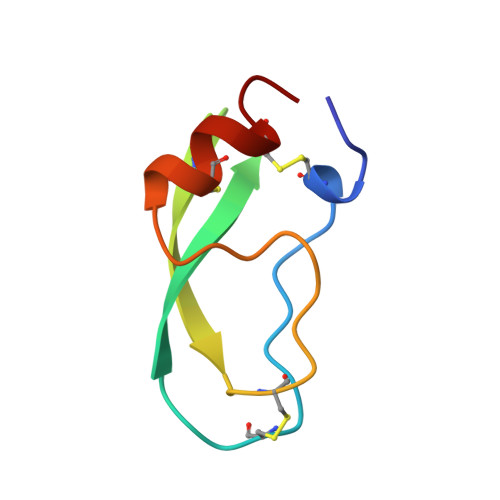
Crystal structure of the bovine alpha-chymotrypsin:Kunitz inhibitor complex. An example of multiple protein:protein recognition sites.
Capasso, C., Rizzi, M., Menegatti, E., Ascenzi, P., Bolognesi, M.(1997) J Mol Recognit 10: 26-35
- PubMed: 9179777
- DOI: https://doi.org/10.1002/(SICI)1099-1352(199701/02)10:1<26::AID-JMR351>3.0.CO;2-N
- Primary Citation of Related Structures:
1MTN - PubMed Abstract:
The crystal structure of bovine alpha-chymotrypsin (alpha-CHT) in complex with the bovine basic pancreatic trypsin inhibitor (BPTI) has been solved and refined at 2.8 A resolution (R-factor = 0.18). The proteinase:inhibitor complex forms a compact dimer (two alpha-CHT and two BPTI molecules), which may be stabilized by surface-bound sulphate ions, in the crystalline state. Each BPTI molecule, at opposite ends, is contacting both proteinase molecules in the dimer, through the reactive site loop and through residues next to the inhibitor's C-terminal region. Specific recognition between alpha-CHT and BPTI occurs at the (re)active site interface according to structural rules inferred from the analysis of homologous serine proteinase:inhibitor complexes. Lys15, the P1 residue of BPTI, however, does not occupy the alpha-CHT S1 specificity pocket, being hydrogen bonded to backbone atoms of the enzyme surface residues Gly216 and Ser217.
- Centro Biotecnologie Avanzate IST, Università di Genova, Italy; C.N.R. Istituto di Biochimica delle Proteine ed Enzimologia, Fuorigrotta Napoli, Italy.
Organizational Affiliation: