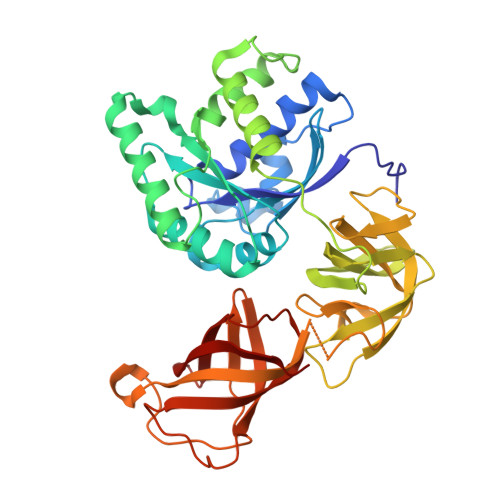
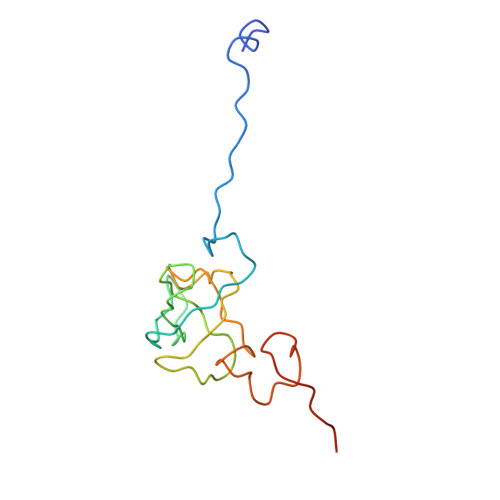
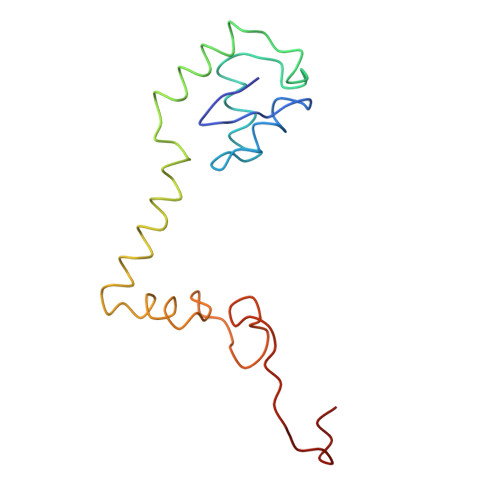
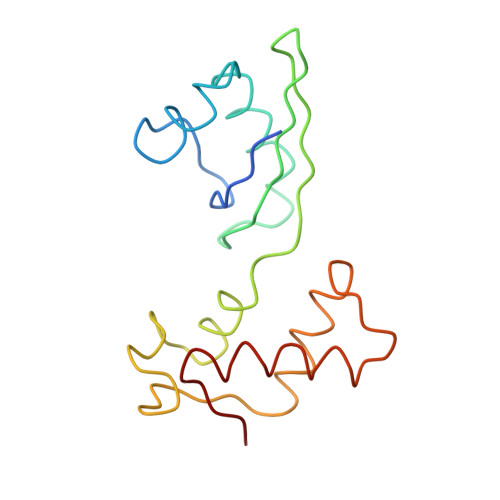
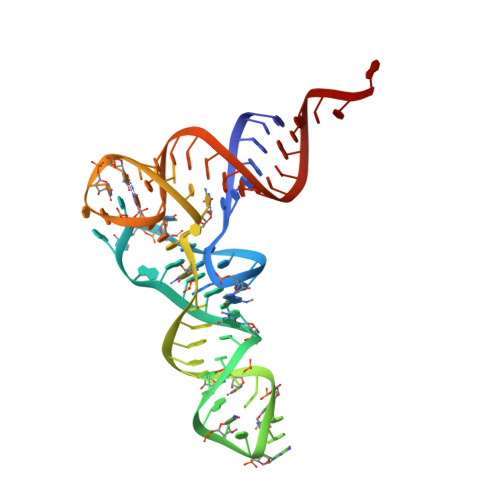
Ribosome Interactions of Aminoacyl-tRNA and Elongation Factor TU in the Codon Recognition Complex
Stark, H., Rodnina, M.V., Wieden, H.-J., Zemlin, F., Wintermeyer, W., van Heel, M.(2002) Nat Struct Biol 9: 849-854
- PubMed: 12379845
- DOI: https://doi.org/10.1038/nsb859
- Primary Citation of Related Structures:
1MJ1 - PubMed Abstract:
The mRNA codon in the ribosomal A-site is recognized by aminoacyl-tRNA (aa-tRNA) in a ternary complex with elongation factor Tu (EF-Tu) and GTP. Here we report the 13 A resolution three-dimensional reconstruction determined by cryo-electron microscopy of the kirromycin-stalled codon-recognition complex. The structure of the ternary complex is distorted by binding of the tRNA anticodon arm in the decoding center. The aa-tRNA interacts with 16S rRNA, helix 69 of 23S rRNA and proteins S12 and L11, while the sarcin-ricin loop of 23S rRNA contacts domain 1 of EF-Tu near the nucleotide-binding pocket. These results provide a detailed snapshot view of an important functional state of the ribosome and suggest mechanisms of decoding and GTPase activation.
- Max-Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany. holger.stark@mpibpc.mpg.de
Organizational Affiliation: