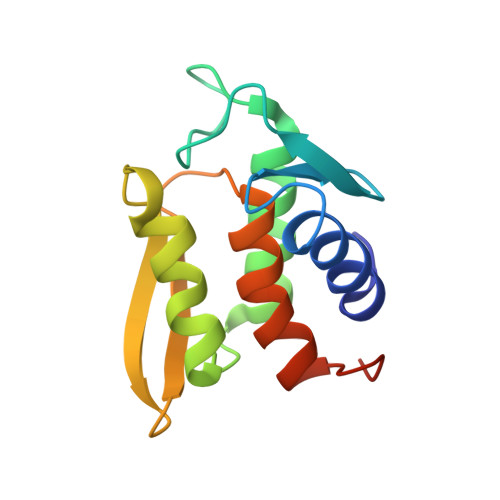
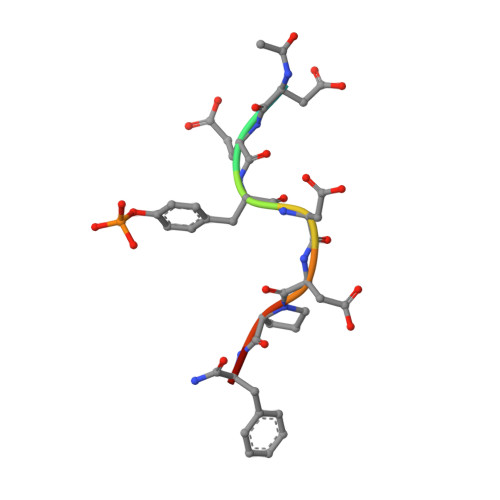
Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Khandelwal, P., Keliikuli, K., Smith, C.L., Saper, M.A., Zuiderweg, E.R.P.(2002) Biochemistry 41: 11425-11437
- PubMed: 12234185
- DOI: https://doi.org/10.1021/bi026333l
- Primary Citation of Related Structures:
1M0V - PubMed Abstract:
Virulence of pathogenic bacteria of the genus Yersinia requires the injection of six effector proteins into the cytoplasm of host cells. The amino-terminal domain of one of these effectors, the tyrosine phosphatase YopH, is essential for translocation of YopH, as well as for targeting it to phosphotyrosine-containing substrates of the type pYxxP. We report the high-resolution solution structure of the N-terminal domain (residues 1-129) from the Yersinia pseudotuberculosis YopH (YopH-NT) in complex with N-acetyl-DEpYDDPF-NH(2), a peptide derived from an in vivo protein substrate. In contrast to the domain-swapped dimer observed in a crystal structure of the same protein (Smith, C. L., Khandelwal, P., Keliikuli, K., Zuiderweg, E. R. P., and Saper, M. A. (2001) Mol. Microbiol. 42, 967-979), YopH-NT is monomeric in solution. The peptide binding site is located on a beta-hairpin that becomes the crossover point in the dimer structure. The binding site has several characteristics that are reminiscent of SH2 domains, which also bind to pYxxP sequences.
- Biophysics Research Division, University of Michigan, 930 North University Avenue, Ann Arbor, Michigan 48109-1055, USA.
Organizational Affiliation: