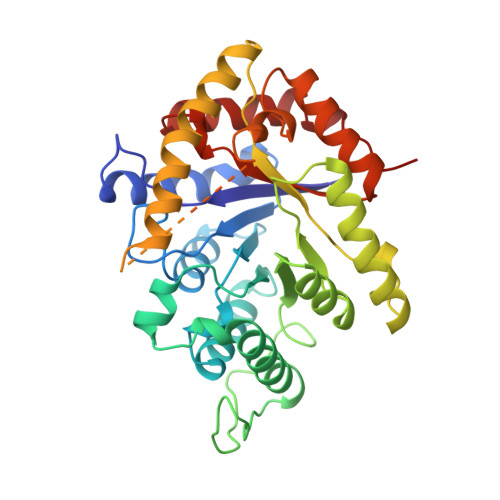
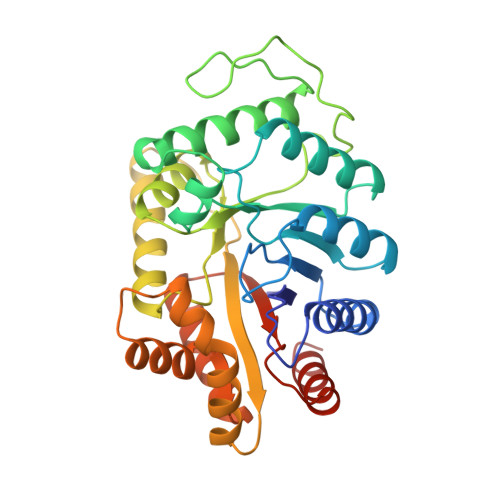
The 1.5-A resolution crystal structure of bacterial luciferase in low salt conditions.
Fisher, A.J., Thompson, T.B., Thoden, J.B., Baldwin, T.O., Rayment, I.(1996) J Biological Chem 271: 21956-21968
- PubMed: 8703001
- DOI: https://doi.org/10.1074/jbc.271.36.21956
- Primary Citation of Related Structures:
1LUC - PubMed Abstract:
Bacterial luciferase is a flavin monooxygenase that catalyzes the oxidation of a long-chain aldehyde and releases energy in the form of visible light. A new crystal form of luciferase cloned from Vibrio harveyi has been grown under low-salt concentrations, which diffract x-rays beyond 1.5-A resolution. The x-ray structure of bacterial luciferase has been refined to a conventional R-factor of 18.2% for all recorded synchrotron data between 30.0 and 1.50-A resolution. Bacterial luciferase is an alpha-beta heterodimer, and the individual subunits fold into a single domain (beta/alpha)8 barrel. The high resolution structure reveals a non-prolyl cis peptide bond that forms between Ala74 and Ala75 in the alpha subunit near the putative active site. This cis peptide bond may have functional significance for creating a cavity at the active site. Bacterial luciferase employs reduced flavin as a substrate rather than a cofactor. The structure presented was determined in the absence of substrates. A comparison of the structural similarities between luciferase and a nonfluorescent flavoprotein, which is expressed in the lux operon of one genus of bioluminescent bacteria, suggests that the two proteins originated from a common ancestor. However, the flavin binding sites of the nonfluorescent protein are likely not representative of the flavin binding site on luciferase. The structure presented here will furnish a detailed molecular model for all bacterial luciferases.
- Institute for Enzyme Research and Department of Biochemistry, University of Wisconsin, Madison, Wisconsin 53705, USA.
Organizational Affiliation: