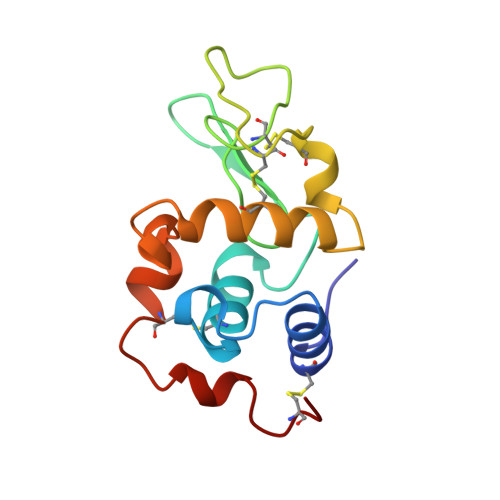
Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme.
Wouters, J.(1998) Protein Sci 7: 2472-2475
- PubMed: 9828016
- DOI: https://doi.org/10.1002/pro.5560071127
- Primary Citation of Related Structures:
1LPI - PubMed Abstract:
Experimental evidence of a cation-pi interaction between a sodium cation (Na+) and the indole ring of residue Trp123 in a structure (2.0 A) of hen egg-white lysozyme is presented. The geometry of the metal ion-pi interaction observed in the protein structure (distance between the aromatic plane and the cation approximately 4 A) is consistent with geometries observed among small molecules crystal structures and quantum chemistry ab initio calculations. The present crystal structure of lysozyme provides unique structural information about the geometry of binding of cations to pi systems in proteins. It shows that the metal ion-pi interaction within proteins is not significantly different from similar bindings found in small molecules and that it can be modeled by theoretical methods.
- Facultés Universitaires N.-D. de la Paix, Namur, Belgium. wouters@scf.fundp.ac.be
Organizational Affiliation: