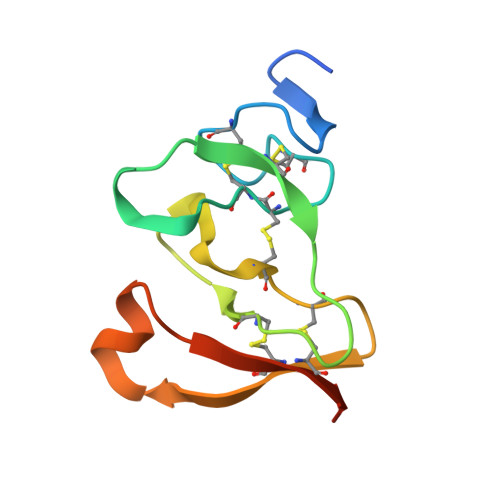
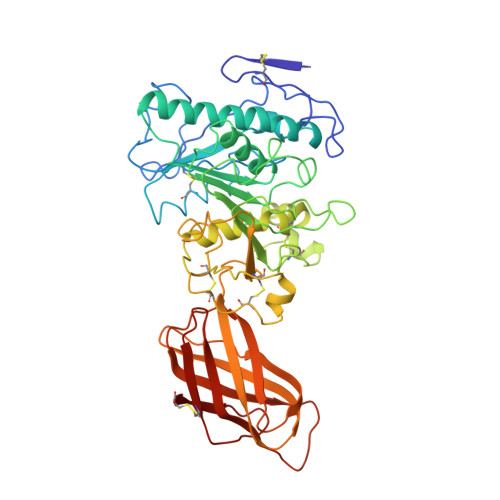
Interfacial activation of the lipase-procolipase complex by mixed micelles revealed by X-ray crystallography.
van Tilbeurgh, H., Egloff, M.P., Martinez, C., Rugani, N., Verger, R., Cambillau, C.(1993) Nature 362: 814-820
- PubMed: 8479519
- DOI: https://doi.org/10.1038/362814a0
- Primary Citation of Related Structures:
1LPA - PubMed Abstract:
The three-dimensional structure of the lipase-procolipase complex, co-crystallized with mixed micelles of phosphatidylcholine and bile salt, has been determined at 3 A resolution by X-ray crystallography. The lid, a surface helix covering the catalytic triad of lipase, adopts a totally different conformation which allows phospholipid to bind to the enzyme's active site. The open lid is an essential component of the active site and interacts with procolipase. Together they form the lipid-water interface binding site. This reorganization of the lid structure provokes a second drastic conformational change in an active site loop, which in its turn creates the oxyanion hole (induced fit).
- LCCMB-CNRS, Faculté de Médecine Nord, Marseilles, France.
Organizational Affiliation: