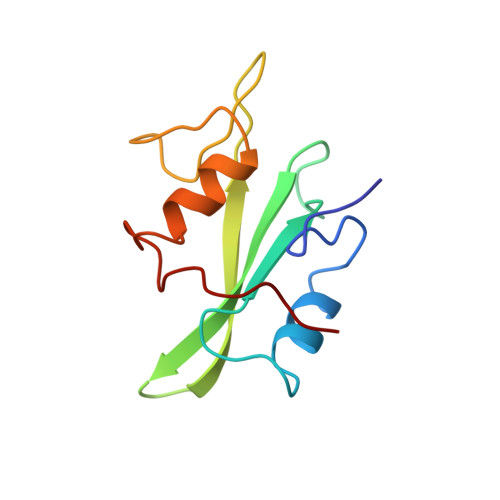
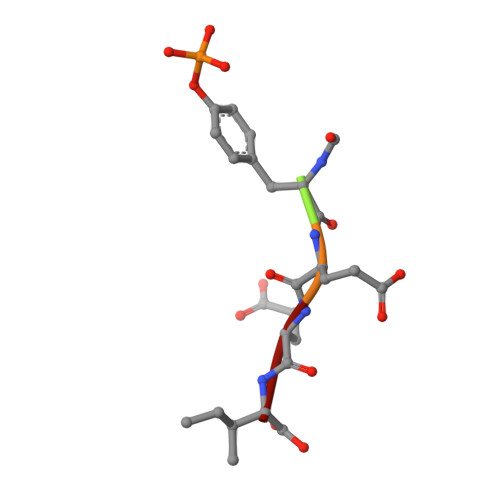
Crystal structures of the human p56lck SH2 domain in complex with two short phosphotyrosyl peptides at 1.0 A and 1.8 A resolution.
Tong, L., Warren, T.C., King, J., Betageri, R., Rose, J., Jakes, S.(1996) J Mol Biology 256: 601-610
- PubMed: 8604142
- DOI: https://doi.org/10.1006/jmbi.1996.0112
- Primary Citation of Related Structures:
1LKK, 1LKL - PubMed Abstract:
src homology 2 (SH2) domains are modules of about 100 amino acid residues and bind to phosphotyrosine-containing motifs in a sequence-specific manner. They play important roles in intracellular signal transduction and represent potential targets for pharmacological intervention. The protein tyrosine kinase p56lck is a member of the src family and is involved in T-cell activation. The crystal structure of its SH2 domain with an 11-residue peptide showed that the phosphotyrosine and the Ile residue at the pY + 3 position are recognized by the SH2 domain. We present here the crystal structure of the SH2 domain of human p56lck in complex with the short phosphotyrosyl peptide Ac-pTyr-Glu-Glu-Ile (pYEEI peptide) at 1.0 A resolution. The structural analysis at atomic resolution reveals that residue Arg134 (alphaA2), which interacts with the phosphotyrosine side-chain, is present in two conformations in the complex. The structure at 1.8 A resolution of the complex with the phosphotyrosyl peptide Ac-pTyr-Glu-Glu-Gly (pYEEG peptide), which is 11 fold less potent, shows another binding mode for the pY + 3 residue as well as rearrangements of the side-chain of Arg196 (EF3) and one of the water molecules at the base of the pY + 3 pocket. The structure of the complex with the short pYEEI peptide at atomic resolution represents a good starting point for the design and optimization of new inhibitors. Comparative structural analysis of many different inhibitor complexes will be an important component of this drug discovery process.
- Department of Inflammatory Diseases, Boehringer Ingelheim Pharmaceuticals Inc. Ridgefield, CT 06877, USA.
Organizational Affiliation: