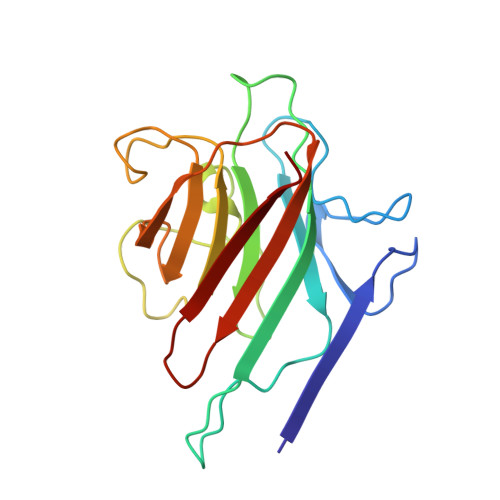
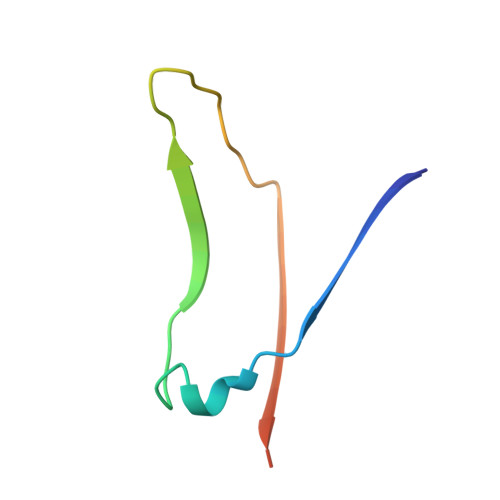
Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Loris, R., Van Overberge, D., Dao-Thi, M.H., Poortmans, F., Maene, N., Wyns, L.(1994) Proteins 20: 330-346
- PubMed: 7731952
- DOI: https://doi.org/10.1002/prot.340200406
- Primary Citation of Related Structures:
1LEN, 2LAL - PubMed Abstract:
The structures of two crystal forms of lentil lectin are determined and refined at high resolution. Orthorhombic lentil lectin is refined at 1.80 A resolution to an R-factor of 0.184 and monoclinic lentil lectin at 1.75 A resolution to an R-factor of 0.175. These two structures are compared to each other and to the other available legume lectin structures. The monosaccharide binding pocket of each lectin monomer contains a tightly bound phosphate ion. This phosphate makes hydrogen bonding contacts with Asp-81 beta, Gly-99 beta, and Asn-125 beta, three residues that are highly conserved in most of the known legume lectin sequences and essential for monosaccharide recognition in all legume lectin crystal structures described thus far. A detailed analysis of the composition and properties of the hydrophobic contact network and hydrophobic nuclei in lentil lectin is presented. Contact map calculations reveal that dense clusters of nonpolar as well as polar side chains play a major role in secondary structure packing. This is illustrated by a large cluster of 24 mainly hydrophobic amino acids that is responsible for the majority of packing interactions between the two beta-sheets. Another series of four smaller and less hydrophobic clusters is found to mediate the packing of a number of loop structures upon the front sheet. A very dense, but not very conserved cluster is found to stabilize the transition metal binding site. The highly conserved and invariant nonpolar residues are distributed asymmetrically over the protein.
- Laboratorium voor Ultrastructuur, Vrije Universiteit Brussel, Sint-Genesius-Rode, Belgium.
Organizational Affiliation: