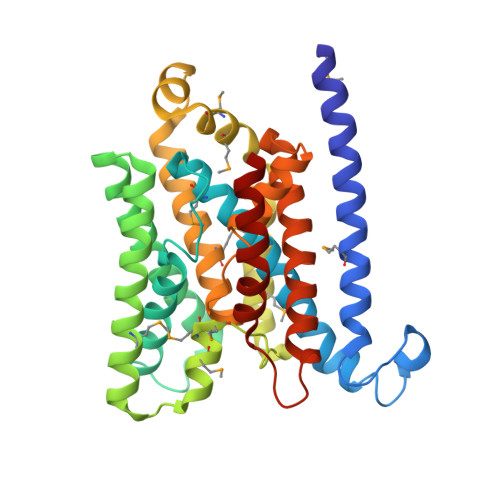
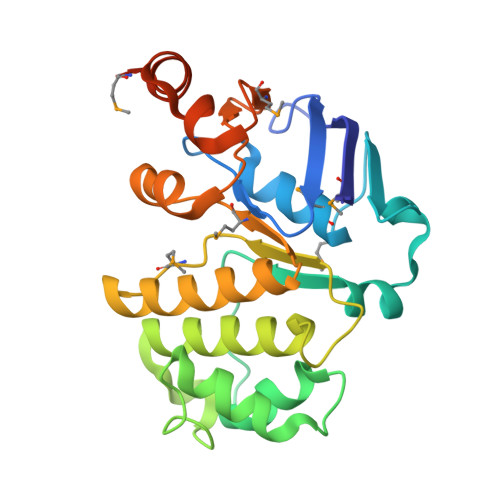
The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism.
Locher, K.P., Lee, A.T., Rees, D.C.(2002) Science 296: 1091-1098
- PubMed: 12004122
- DOI: https://doi.org/10.1126/science.1071142
- Primary Citation of Related Structures:
1L7V - PubMed Abstract:
The ABC transporters are ubiquitous membrane proteins that couple adenosine triphosphate (ATP) hydrolysis to the translocation of diverse substrates across cell membranes. Clinically relevant examples are associated with cystic fibrosis and with multidrug resistance of pathogenic bacteria and cancer cells. Here, we report the crystal structure at 3.2 angstrom resolution of the Escherichia coli BtuCD protein, an ABC transporter mediating vitamin B12 uptake. The two ATP-binding cassettes (BtuD) are in close contact with each other, as are the two membrane-spanning subunits (BtuC); this arrangement is distinct from that observed for the E. coli lipid flippase MsbA. The BtuC subunits provide 20 transmembrane helices grouped around a translocation pathway that is closed to the cytoplasm by a gate region whereas the dimer arrangement of the BtuD subunits resembles the ATP-bound form of the Rad50 DNA repair enzyme. A prominent cytoplasmic loop of BtuC forms the contact region with the ATP-binding cassette and appears to represent a conserved motif among the ABC transporters.
- Howard Hughes Medical Institute and Division of Chemistry and Chemical Engineering, Mail Code 147-75CH, California Institute of Technology, Pasadena, CA 91125, USA. locher@caltech.edu
Organizational Affiliation: