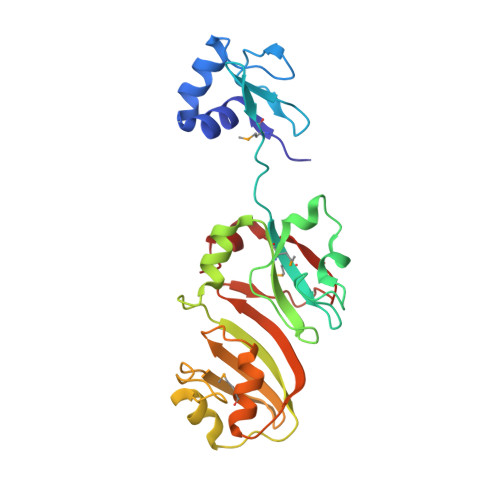
Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Sivaraman, J., Sauve, V., Larocque, R., Stura, E.A., Schrag, J.D., Cygler, M., Matte, A.(2002) Nat Struct Biol 9: 353-358
- PubMed: 11953756
- DOI: https://doi.org/10.1038/nsb788
- Primary Citation of Related Structures:
1KSK, 1KSL, 1KSV - PubMed Abstract:
In Escherichia coli, the pseudouridine synthase RsuA catalyzes formation of pseudouridine (psi) at position 516 in 16S rRNA during assembly of the 30S ribosomal subunit. We have determined the crystal structure of RsuA bound to uracil at 2.0 A resolution and to uridine 5'-monophosphate (UMP) at 2.65 A resolution. RsuA consists of an N-terminal domain connected by an extended linker to the central and C-terminal domains. Uracil and UMP bind in a cleft between the central and C-terminal domains near the catalytic residue Asp 102. The N-terminal domain shows structural similarity to the ribosomal protein S4. Despite only 15% amino acid identity, the other two domains are structurally similar to those of the tRNA-specific psi-synthase TruA, including the position of the catalytic Asp. Our results suggest that all four families of pseudouridine synthases share the same fold of their catalytic domain(s) and uracil-binding site.
- Biotechnology Research Institute, 6100 Royalmount Avenue, Montreal, Quebec H4P 2R2, Canada.
Organizational Affiliation: