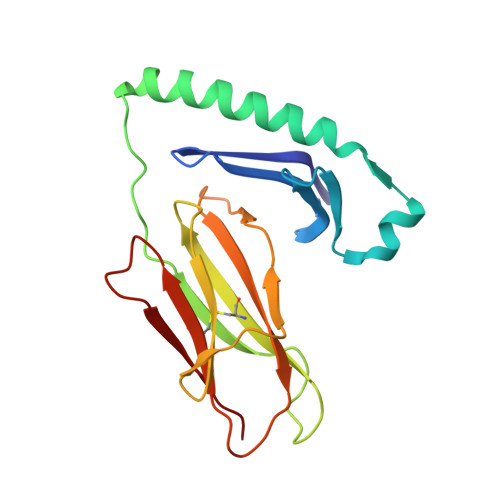
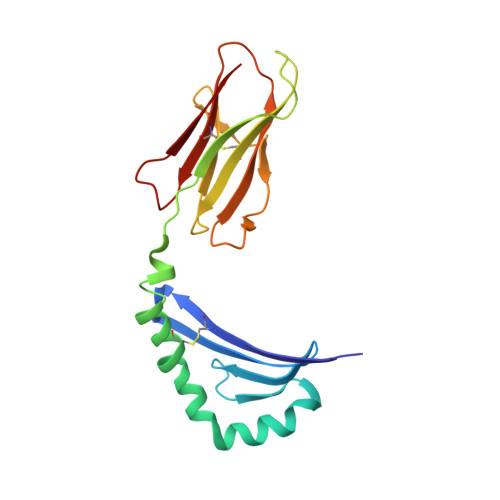
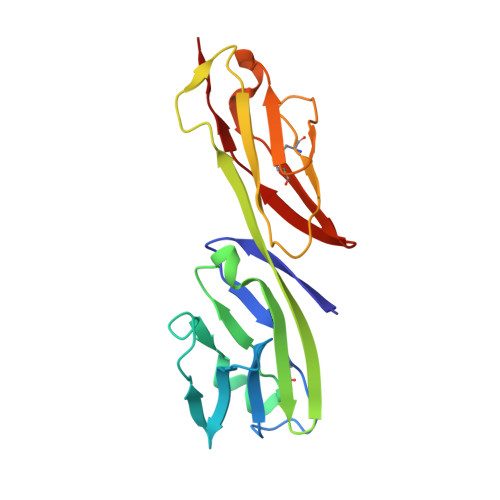
Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Wang, J.H., Meijers, R., Xiong, Y., Liu, J.H., Sakihama, T., Zhang, R., Joachimiak, A., Reinherz, E.L.(2001) Proc Natl Acad Sci U S A 98: 10799-10804
- PubMed: 11535811
- DOI: https://doi.org/10.1073/pnas.191124098
- Primary Citation of Related Structures:
1JL4 - PubMed Abstract:
The structural basis of the interaction between the CD4 coreceptor and a class II major histocompatibility complex (MHC) is described. The crystal structure of a complex containing the human CD4 N-terminal two-domain fragment and the murine I-A(k) class II MHC molecule with associated peptide (pMHCII) shows that only the "top corner" of the CD4 molecule directly contacts pMHCII. The CD4 Phe-43 side chain extends into a hydrophobic concavity formed by MHC residues from both alpha 2 and beta 2 domains. A ternary model of the CD4-pMHCII-T-cell receptor (TCR) reveals that the complex appears V-shaped with the membrane-proximal pMHCII at the apex. This configuration excludes a direct TCR-CD4 interaction and suggests how TCR and CD4 signaling is coordinated around the antigenic pMHCII complex. Human CD4 binds to HIV gp120 in a manner strikingly similar to the way in which CD4 interacts with pMHCII. Additional contacts between gp120 and CD4 give the CD4-gp120 complex a greater affinity. Thus, ligation of the viral envelope glycoprotein to CD4 occludes the pMHCII-binding site on CD4, contributing to immunodeficiency.
- Laboratory of Immunobiology and Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA. jwang@dfci.harvard.edu
Organizational Affiliation: