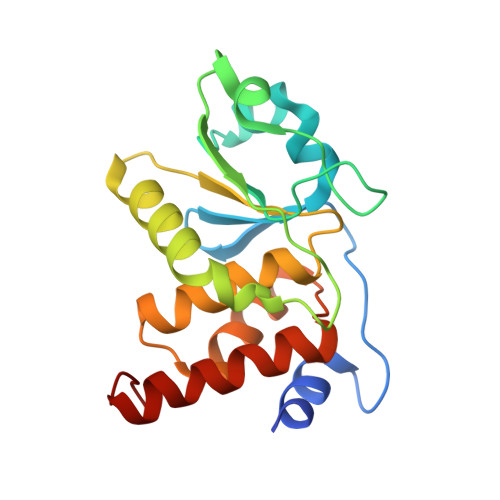
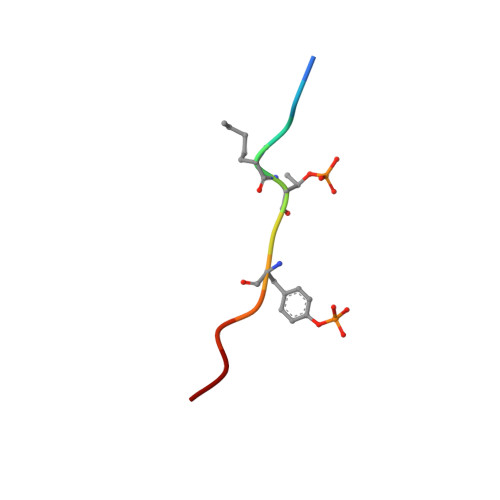
Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein Phosphatase.
Schumacher, M.A., Todd, J.L., Rice, A.E., Tanner, K.G., Denu, J.M.(2002) Biochemistry 41: 3009-3017
- PubMed: 11863439
- DOI: https://doi.org/10.1021/bi015799l
- Primary Citation of Related Structures:
1J4X - PubMed Abstract:
Human VHR (vaccinia H1 related phosphatase) is a member of the dual-specificity phosphatases (DSPs) that often act on bisphosphorylated protein substrates. Unlike most DSPs, VHR displays a strong preference for dephosphorylating phosphotyrosine residues over phosphothreonine residues. Here we describe the 2.75 A crystal structure of the C124S inactive VHR mutant in complex with a bisphosphorylated peptide corresponding to the MAP kinase activation lip. This structure and subsequent biochemical studies revealed the basis for the strong preference for hydrolyzing phosphotyrosine within bisphosphorylated substrates containing -pTXpY-. In the structure, the two phospho residues are oriented into distinct pockets; the phosphotyrosine is bound in the exposed yet deep active site cleft while the phosphothreonine is loosely tethered into a nearby basic pocket containing Arg(158). As this structure is the first substrate-enzyme complex reported for the DSP family of enzymes, these results provide the first glimpse into how DSPs bind their protein substrates.
- Department of Biochemistry and Molecular Biology, Oregon Health and Science University, Portland, Oregon 97201-3098, USA. schumacn@ohsu.edu
Organizational Affiliation: