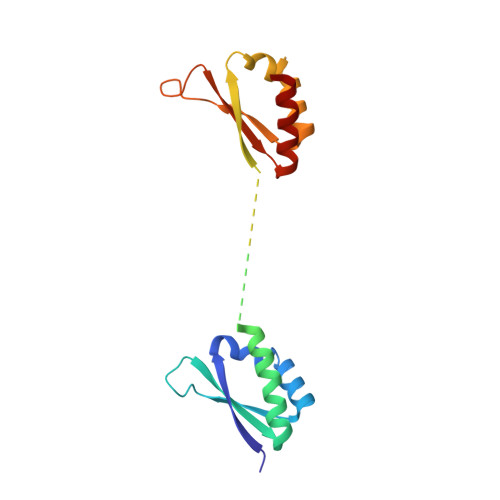
Structure and dynamics of KH domains from FBP bound to single-stranded DNA.
Braddock, D.T., Louis, J.M., Baber, J.L., Levens, D., Clore, G.M.(2002) Nature 415: 1051-1056
- PubMed: 11875576
- DOI: https://doi.org/10.1038/4151051a
- Primary Citation of Related Structures:
1J4W - PubMed Abstract:
Gene regulation can be tightly controlled by recognition of DNA deformations that are induced by stress generated during transcription. The KH domains of the FUSE-binding protein (FBP), a regulator of c-myc expression, bind in vivo and in vitro to the single-stranded far-upstream element (FUSE), 1,500 base pairs upstream from the c-myc promoter. FBP bound to FUSE acts through TFIIH at the promoter. Here we report the solution structure of a complex between the KH3 and KH4 domains of FBP and a 29-base single-stranded DNA from FUSE. The KH domains recognize two sites, 9-10 bases in length, separated by 5 bases, with KH4 bound to the 5' site and KH3 to the 3' site. The central portion of each site comprises a tetrad of sequence 5'd-ATTC for KH4 and 5'd-TTTT for KH3. Dynamics measurements show that the two KH domains bind as articulated modules to single-stranded DNA, providing a flexible framework with which to recognize transient, moving targets.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: