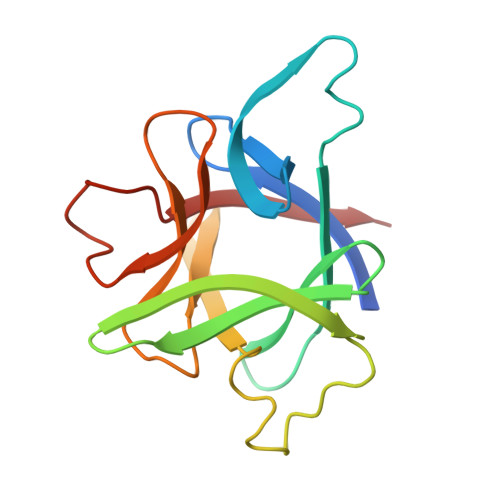
X-ray structure of interleukin-1 receptor antagonist at 2.0-A resolution.
Vigers, G.P., Caffes, P., Evans, R.J., Thompson, R.C., Eisenberg, S.P., Brandhuber, B.J.(1994) J Biological Chem 269: 12874-12879
- PubMed: 8175703
- Primary Citation of Related Structures:
1ILT - PubMed Abstract:
Interleukin-1 receptor antagonist (IL-1ra) is a natural competitive antagonist of IL-1. In order to further elucidate the mechanism by which IL-1ra binds without activating the IL-1 receptor, we have solved the crystal structure of IL-1ra at 2.0-A resolution. IL-1ra has the same overall beta-trefoil fold as IL-1 alpha and IL-1 beta and has a very similar hydrophobic core. However, there are a number of structural differences between the molecules, including significant differences at the open end of the beta-barrel, which has been identified in IL-1 beta as a receptor binding site.
- Synergen, Inc., Boulder, Colorado 80301.
Organizational Affiliation: