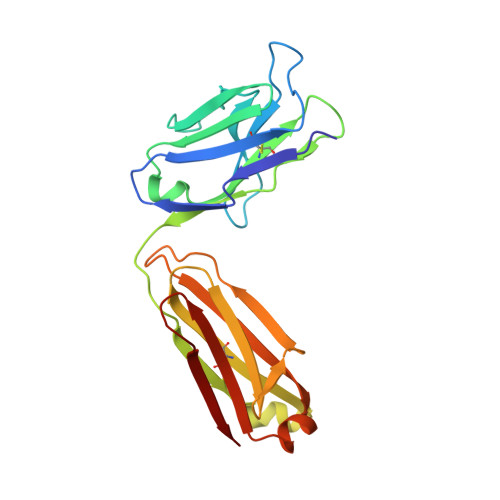
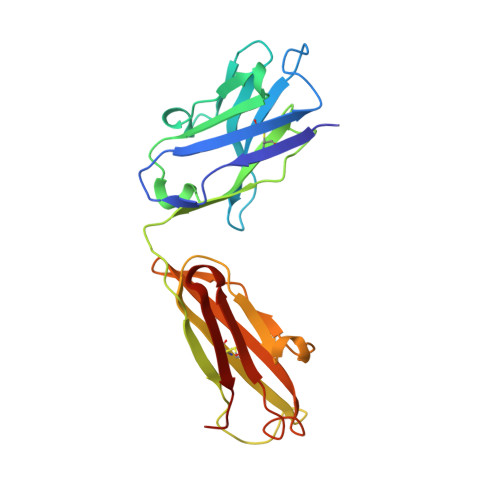
Major antigen-induced domain rearrangements in an antibody.
Stanfield, R.L., Takimoto-Kamimura, M., Rini, J.M., Profy, A.T., Wilson, I.A.(1993) Structure 1: 83-93
- PubMed: 8069628
- DOI: https://doi.org/10.1016/0969-2126(93)90024-b
- Primary Citation of Related Structures:
1GGB, 1GGC - PubMed Abstract:
Recent structural results have shown that antibodies use an induced fit mechanism to recognize and bind their antigens. Here we present the crystallographically determined structure of an Fab directed against an HIV-1 peptide (Fab 50.1) in the unliganded state and compare it with the peptide-bound structure. We perform a detailed analysis of the components that contribute to enhanced antigen binding and recognition. Induced fit of Fab 50.1 to its peptide antigen involves a substantial rearrangement of the third complementarity determining region loop of the heavy chain (H3), as well as a large rotation of the variable heavy (VH) chain relative to the variable light (VL) chain. Analysis of other Fab structures suggests that the extent of the surface area buried at the VL-VH interface correlates with the ability to alter antibody quaternary structure by reorientation of the VL-VH domains. Fab 50.1 exhibits the largest conformational changes yet observed in a single antibody. These can be attributed to the flexibility of the variable region. Comparisons of new data with previous examples lend to the general conclusion that a small VL-VH interface, due in part to a short H3 loop, permits substantial alterations to the antigen-binding pocket. This has major implications for the prediction, engineering and design of antibody-combining sites.
- Department of Molecular Biology, Scripps Research Institute, La Jolla, CA 92037.
Organizational Affiliation: