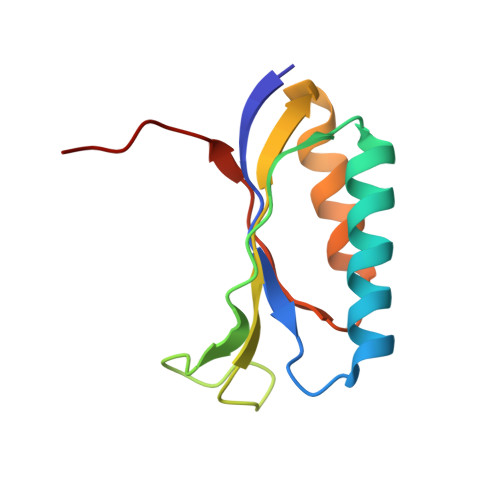
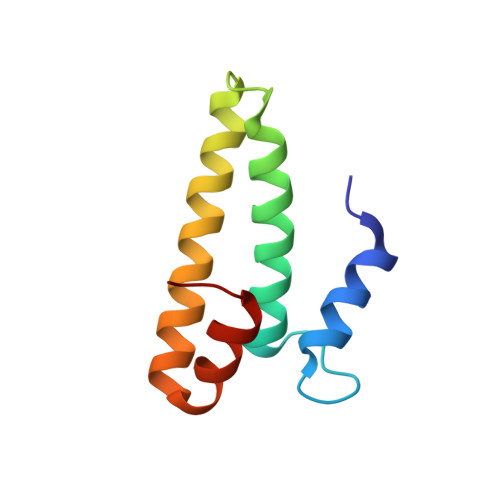
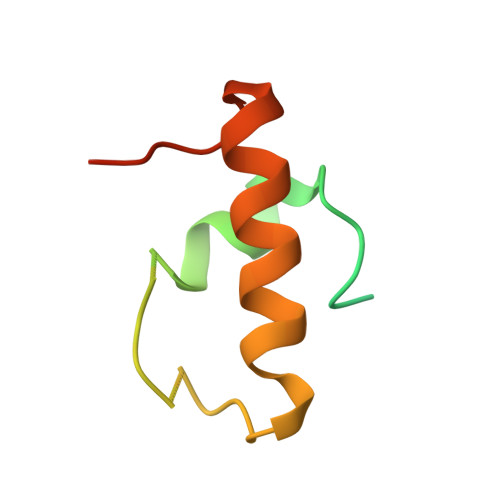
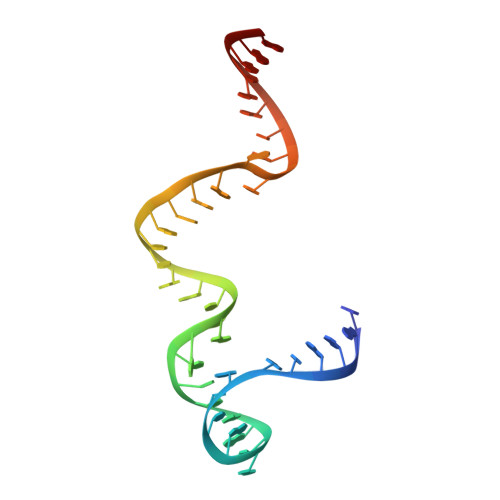
Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain.
Agalarov, S.C., Sridhar Prasad, G., Funke, P.M., Stout, C.D., Williamson, J.R.(2000) Science 288: 107-112
- PubMed: 10753109
- DOI: https://doi.org/10.1126/science.288.5463.107
- Primary Citation of Related Structures:
1G1X - PubMed Abstract:
The crystal structure of a 70-kilodalton ribonucleoprotein complex from the central domain of the Thermus thermophilus 30S ribosomal subunit was solved at 2.6 angstrom resolution. The complex consists of a 104-nucleotide RNA fragment composed of two three-helix junctions that lie at the end of a central helix, and the ribosomal proteins S15, S6, and S18. S15 binds the ribosomal RNA early in the assembly of the 30S ribosomal subunit, stabilizing a conformational reorganization of the two three-helix junctions that creates the RNA fold necessary for subsequent binding of S6 and S18. The structure of the complex demonstrates the central role of S15-induced reorganization of central domain RNA for the subsequent steps of ribosome assembly.
- Department of Molecular Biology and the Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA 92037, USA. dave@scripps.edu
Organizational Affiliation: