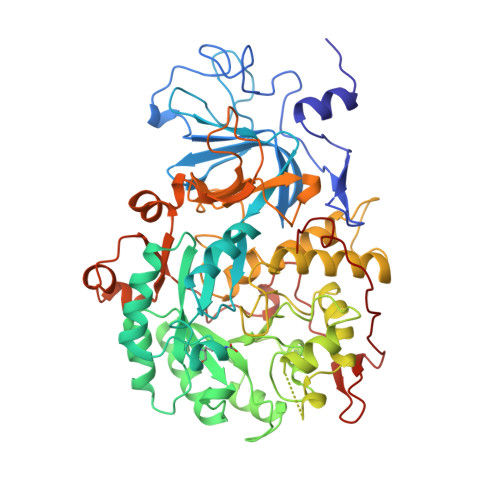
Characterization of the mononickel metallocenter in H134A mutant urease.
Park, I.S., Michel, L.O., Pearson, M.A., Jabri, E., Karplus, P.A., Wang, S., Dong, J., Scott, R.A., Koehler, B.P., Johnson, M.K., Hausinger, R.P.(1996) J Biological Chem 271: 18632-18637
- PubMed: 8702515
- DOI: https://doi.org/10.1074/jbc.271.31.18632
- Primary Citation of Related Structures:
1FWI - PubMed Abstract:
A mutant form of Klebsiella aerogenes urease possessing Ala instead of His at position 134 (H134A) is inactive and binds approximately half the normal complement of nickel (Park, I.-S., and Hausinger, R. P.(1993) Protein Sci. 2, 1034-1041). The crystal structure of the H134A protein was obtained at 2.0-A resolution, and it confirms that only Ni-1 of the two nickel ions found in the native enzyme is present. In contrast to the pseudotetrahedral geometry observed for Ni-1 in native urease (where it is liganded by His-246, His-272, one oxygen atom of carbamylated Lys-217, and a water molecule at partial occupancy), the mononickel metallocenter in the H134A protein was found to possess octahedral geometry and was coordinated by the above protein ligands plus three water molecules. The nickel site of H134A urease was probed by UV-visible, variable temperature magnetic circular dichroism, and x-ray absorption spectroscopies. The spectroscopic data are consistent with the presence of Ni(II) in octahedral geometry coordinated by two histidylimidazoles and additional oxygen and/or nitrogen donors. These data underscore the requirement of Ni-2 for formation of active urease and demonstrate the important role of Ni-2 in establishing the proper Ni-1 coordination geometry.
- Department of Microbiology, Michigan State University, East Lansing, Michigan 48824-1101, USA.
Organizational Affiliation: