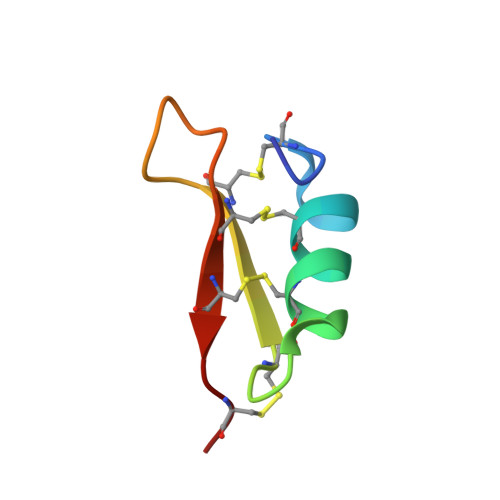
Solution structure and activity of the synthetic four-disulfide bond Mediterranean mussel defensin (MGD-1).
Yang, Y.S., Mitta, G., Chavanieu, A., Calas, B., Sanchez, J.F., Roch, P., Aumelas, A.(2000) Biochemistry 39: 14436-14447
- PubMed: 11087396
- DOI: https://doi.org/10.1021/bi0011835
- Primary Citation of Related Structures:
1FJN - PubMed Abstract:
MGD-1 is a 39-residue defensin-like peptide isolated from the edible Mediterranean mussel, Mytilus galloprovincialis. This peptide is characterized by the presence of four disulfide bonds. We report here its solid-phase synthesis and an easy way to improve the yield of the four native disulfide bonds. Synthetic and native MGD-1 display similar antibacterial activity, suggesting that the hydroxylation of Trp28 observed in native MGD-1 is not involved in the antimicrobial effect. The three-dimensional solution structure of MGD-1 has been established using (1)H NMR and mainly consists of a helical part (Asn7-Ser16) and two antiparallel beta-strands (Arg20-Cys25 and Cys33-Arg37), together giving rise to the common cystine-stabilized alpha-beta motif frequently observed in scorpion toxins. In MGD-1, the cystine-stabilized alpha-beta motif is stabilized by four disulfide bonds (Cys4-Cys25, Cys10-Cys33, Cys14-Cys35, and Cys21-Cys38), instead of by the three disulfide bonds commonly found in arthropod defensins. Except for the Cys21-Cys38 disulfide bond which is solvent-exposed, the three others belong to the particularly hydrophobic core of the highly constrained structure. Moreover, the C4-P5 amide bond in the cis conformation characterizes the MGD-1 structure. MGD-1 and insect defensin A possess similar bactericidal anti-Gram-positive activity, suggesting that the fourth disulfide bond of MGD-1 is not essential for the biological activity. In agreement with the general features of antibacterial peptides, the MGD-1 and defensin A structures display a typical distribution of positively charged and hydrophobic side chains. The positively charged residues of MGD-1 are located in three clusters. For these two defensin peptides isolated from insects and mollusks, it appears that the rather well conserved location of certain positively charged residues and of the large hydrophobic cluster are enough to generate the bactericidal potency and the Gram-positive specificity.
- Centre de Biochimie Structurale, CNRS UMR 5048, INSERM U414, Université Montpellier 1, France.
Organizational Affiliation: