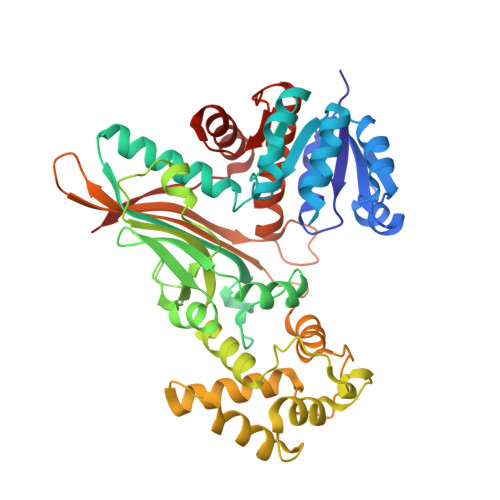
Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the alpha-aminoadipate pathway of lysine biosynthesis.
Johansson, E., Steffens, J.J., Lindqvist, Y., Schneider, G.(2000) Structure 8: 1037-1047
- PubMed: 11080625
- DOI: https://doi.org/10.1016/s0969-2126(00)00512-8
- Primary Citation of Related Structures:
1E5L, 1E5Q, 1FF9 - PubMed Abstract:
The biosynthesis of the essential amino acid lysine in higher fungi and cyanobacteria occurs via the alpha-aminoadipate pathway, which is completely different from the lysine biosynthetic pathway found in plants and bacteria. The penultimate reaction in the alpha-aminoadipate pathway is catalysed by NADPH-dependent saccharopine reductase. We set out to determine the structure of this enzyme as a first step in exploring the structural biology of fungal lysine biosynthesis. We have determined the three-dimensional structure of saccharopine reductase from the plant pathogen Magnaporthe grisea in its apo form to 2.0 A resolution and as a ternary complex with NADPH and saccharopine to 2.1 A resolution. Saccharopine reductase is a homodimer, and each subunit consists of three domains, which are not consecutive in amino acid sequence. Domain I contains a variant of the Rossmann fold that binds NADPH. Domain II folds into a mixed seven-stranded beta sheet flanked by alpha helices and is involved in substrate binding and dimer formation. Domain III is all-helical. The structure analysis of the ternary complex reveals a large movement of domain III upon ligand binding. The active site is positioned in a cleft between the NADPH-binding domain and the second alpha/beta domain. Saccharopine is tightly bound to the enzyme via a number of hydrogen bonds to invariant amino acid residues. On the basis of the structure of the ternary complex of saccharopine reductase, an enzymatic mechanism is proposed that includes the formation of a Schiff base as a key intermediate. Despite the lack of overall sequence homology, the fold of saccharopine reductase is similar to that observed in some enzymes of the diaminopimelate pathway of lysine biosynthesis in bacteria. These structural similarities suggest an evolutionary relationship between two different major families of amino acid biosynthetic pathway, the glutamate and aspartate families.
- Department of Medical Biochemistry and Biophysics Karolinska Institutet S-171 77, Stockholm, Sweden.
Organizational Affiliation: