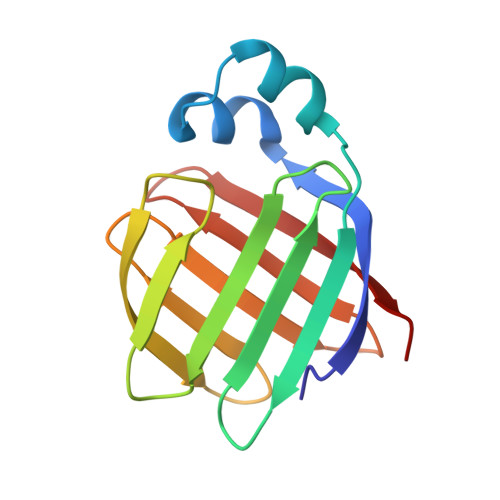
Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Lucke, C., Zhang, F., Ruterjans, H., Hamilton, J.A., Sacchettini, J.C.(1996) Structure 4: 785-800
- PubMed: 8805562
- DOI: https://doi.org/10.1016/s0969-2126(96)00086-x
- Primary Citation of Related Structures:
1EAL - PubMed Abstract:
The family of lipid binding proteins (LBPs) includes a large number of fatty acid binding proteins (FABPs) but only two proteins (ileal lipid binding protein, ILBP, and liver fatty acid binding protein) that can bind both fatty acids and bile acids. Bile acid transport is medically and pharmacologically important, but is poorly understood. To understand the binding properties of ILBP, we studied its solution structure with and without bound lipids and compared these with known structures of FABPs. The sequence-specific 1H resonance assignments for porcine ILBP have been determined by homonuclear two-dimensional (2D) NMR spectroscopy for the apo-protein as well as for ILBP complexes with fatty acid and bile acid ligands. From NOE spectra and hydrogen exchange data, similar secondary structure elements were identified for all three protein forms. ILBP is composed of ten antiparallel beta strands arranged in two nearly orthogonal beta sheets (a fold seen in other FABPs, and dubbed the "beta-clam shell'), covered on one side by two short, nearly parallel alpha helices. Binding of fatty acids or bile acids to ILBP alters mainly the side-chain proton resonances of amino acids within the protein cavity, indicating that both bile acids and fatty acids can bind in the interior of the protein between the two beta sheets; binding of bile acids stabilizes the protein backbone by a small amount. Fast hydrogen exchange rates for the backbone amide protons of ILBP indicate that the hydrogen-bonding network of the beta sheet in ILBP is weaker than the corresponding network in rat intestinal and bovine heart FABPs. The tertiary structure of ILBP is similar to that of other LBPs, but appears to be unusually flexible, with a relatively weak hydrogen-bonding network. It is likely that this flexibility is important in allowing bile acids, which are larger and more rigid than fatty acids, to enter the central cavity of the protein.
- Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Organizational Affiliation: