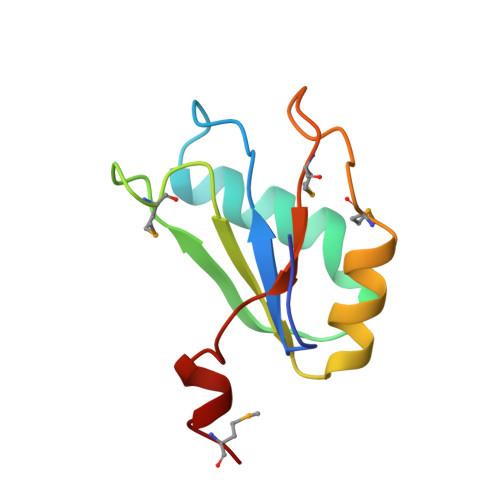
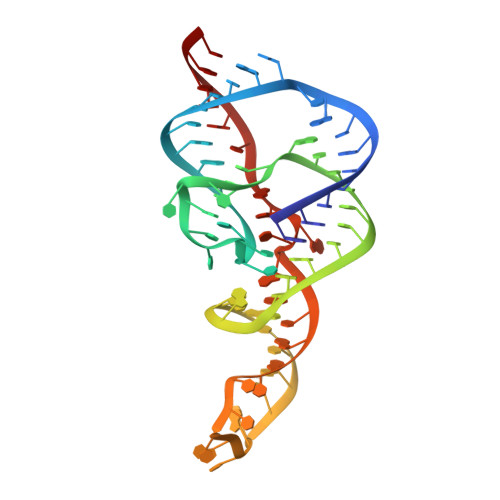
Crystal structure of a hepatitis delta virus ribozyme.
Ferre-D'Amare, A.R., Zhou, K., Doudna, J.A.(1998) Nature 395: 567-574
- PubMed: 9783582
- DOI: https://doi.org/10.1038/26912
- Primary Citation of Related Structures:
1DRZ - PubMed Abstract:
The self-cleaving ribozyme of the hepatitis delta virus (HDV) is the only catalytic RNA known to be required for the viability of a human pathogen. We obtained crystals of a 72-nucleotide, self-cleaved form of the genomic HDV ribozyme that diffract X-rays to 2.3 A resolution by engineering the RNA to bind a small, basic protein without affecting ribozyme activity. The co-crystal structure shows that the compact catalytic core comprises five helical segments connected as an intricate nested double pseudoknot. The 5'-hydroxyl leaving group resulting from the self-scission reaction is buried deep within an active-site cleft produced by juxtaposition of the helices and five strand-crossovers, and is surrounded by biochemically important backbone and base functional groups in a manner reminiscent of protein enzymes.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520-8114, USA.
Organizational Affiliation: