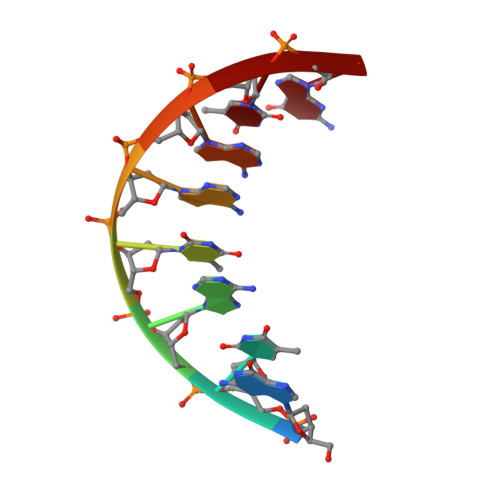
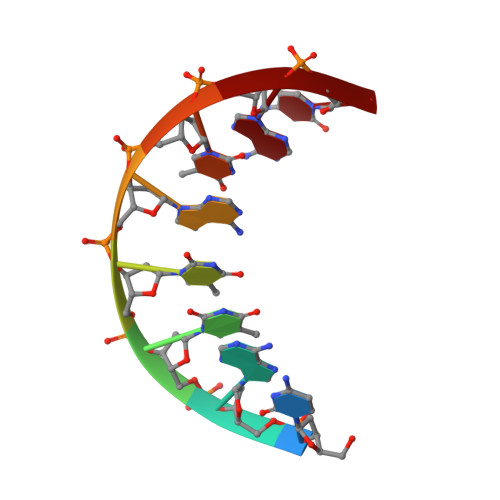
Solution structure of a DNA octamer containing the Pribnow box via restrained molecular dynamics simulation with distance and torsion angle constraints derived from two-dimensional nuclear magnetic resonance spectral fitting.
Schmitz, U., Sethson, I., Egan, W.M., James, T.L.(1992) J Mol Biology 227: 510-531
- PubMed: 1404366
- DOI: https://doi.org/10.1016/0022-2836(92)90904-x
- Primary Citation of Related Structures:
1D70 - PubMed Abstract:
The DNA octamer [d(GTATAATG].[(CATATTAC)], containing the prokaryotic upstream consensus recognition sequence, has been examined via proton homonuclear two-dimensional nuclear Overhauser effect (2D NOE) and double-quantum-filtered correlation (2QF-COSY) spectra. All proton resonances, except those of H5' and H5" protons, were assigned. A temperature dependence study of one-dimensional nuclear magnetic resonance (NMR) spectra, rotating frame 2D NOE spectroscopy (ROESY), and T1 rho measurements revealed an exchange process that apparently is global in scope. Work at lower temperatures enabled a determination of structural constraints that could be employed in determination of a time-averaged structure. Simulations of the 2QF-COSY cross-peaks were compared with experimental data, establishing scalar coupling constant ranges of the individual sugar ring protons and hence pucker parameters for individual deoxyribose rings. The rings exhibit a dynamic equilibrium of N and S-type conformers with 80 to 100% populations of the latter. A program for iterative complete relaxation matrix analysis of 2D NOE spectral intensities, MARDIGRAS, was employed to give interproton distances for each mixing time. According to the accuracy of the distance determination, upper and lower distance bounds were chosen. The distance bounds define the size of a flat-well potential function term, incorporated into the AMBER force-field, which was employed for restrained molecular dynamics calculations. Torsion angle constraints in the form of a flat-well potential were also constructed from the analysis of the sugar pucker data. Several restrained molecular dynamics runs of 25 picoseconds were performed, utilizing 184 experimental distance constraints and 80 torsion angle constraints; three different starting structures were used: energy minimized A-DNA, B-DNA, and wrinkled D-DNA, another member of the B-DNA family. Convergence to similar structures obtained with root-mean-square deviations between resulting structures of 0.37 to 0.92 A for the central hexamer of the octamer. The average structure from the nine different molecular dynamics runs was subjected to final restrained energy minimization. The resulting final structure was in good agreement with the structures derived from different molecular dynamics runs and exhibited a substantial improvement in the 2D NOE sixth-root residual index in comparison with the starting structures. An approximation of the structure in the terminal base-pairs, which displayed experimental evidence of fraying, was made by maintaining the structure of the inner four base-pairs and performing molecular dynamics simulations with the experimental structural constraints observed for the termini.(ABSTRACT TRUNCATED AT 400 WORDS)
- Department of Pharmaceutical Chemistry, University of California, San Francisco 94143-0446.
Organizational Affiliation: