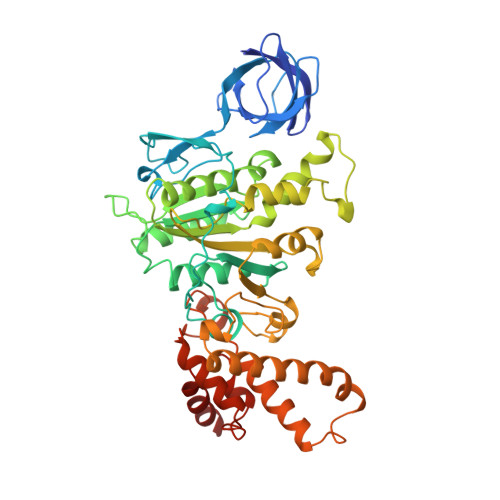
The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B.
van Raaij, M.J., Abrahams, J.P., Leslie, A.G., Walker, J.E.(1996) Proc Natl Acad Sci U S A 93: 6913-6917
- PubMed: 8692918
- DOI: https://doi.org/10.1073/pnas.93.14.6913
- Primary Citation of Related Structures:
1COW - PubMed Abstract:
In the structure of bovine mitochondrial F1-ATPase that was previously determined with crystals grown in the presence of adenylyl-imidodiphosphate (AMP-PNP) and ADP, the three catalytic beta-subunits have different conformations and nucleotide occupancies. Adenylyl-imidodiphosphate is bound to one beta-subunit (betaTP), ADP is bound to the second (betaDP), and no nucleotide is bound to the third (betaE). Here we show that the uncompetitive inhibitor aurovertin B binds to bovine F1 at two equivalent sites in betaTP and betaE, in a cleft between the nucleotide binding and C-terminal domains. In betaDP, the aurovertin B pocket is incomplete and is inaccessible to the inhibitor. The aurovertin B bound to betaTP interacts with alpha-Glu399 in the adjacent alphaTP subunit, whereas the aurovertin B bound to betaE is too distant from alphaE to make an equivalent interaction. Both sites encompass betaArg-412, which was shown by mutational studies to be involved in binding aurovertin. Except for minor changes around the aurovertin pockets, the structure of bovine F1-ATPase is the same as determined previously. Aurovertin B appears to act by preventing closure of the catalytic interfaces, which is essential for a catalytic mechanism involving cyclic interconversion of catalytic sites.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: