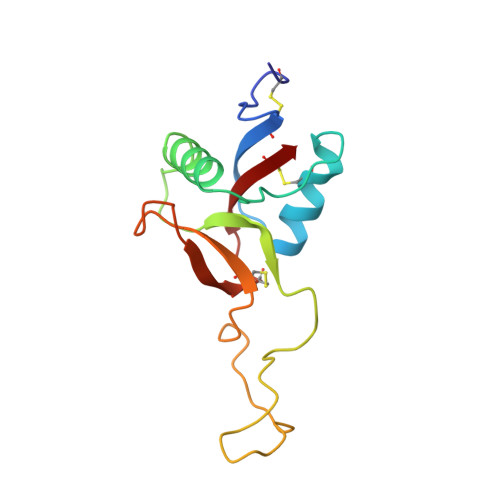
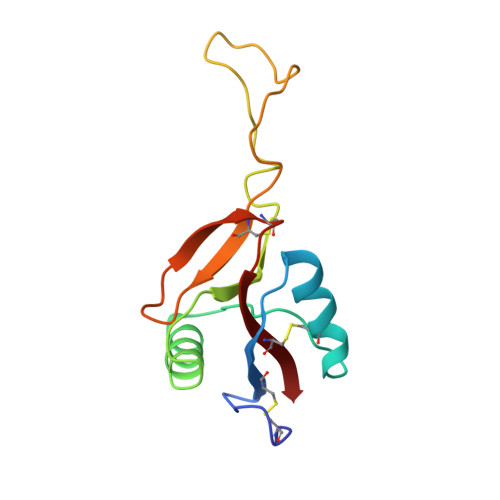
Crystal structure of coagulation factor IX-binding protein from habu snake venom at 2.6 A: implication of central loop swapping based on deletion in the linker region.
Mizuno, H., Fujimoto, Z., Koizumi, M., Kano, H., Atoda, H., Morita, T.(1999) J Mol Biology 289: 103-112
- PubMed: 10339409
- DOI: https://doi.org/10.1006/jmbi.1999.2756
- Primary Citation of Related Structures:
1BJ3 - PubMed Abstract:
Coagulation factor IX-binding protein (IX-bp) isolated from the venom of the habu snake (Trimeresurus flavoviridis) is a disulfide-linked heterodimer consisting of homologous subunits A and B. The structure of IX-bp has been solved by X-ray crystallography at 2.6 A resolution to a crystallographic R -value of 0.181. The main-chain fold of each subunit is homologous to the carbohydrate-recognition domain of C-type lectins (C-type CRDs) except for the extended central loop. The structure is almost identical with that of factors IX and X-binding protein (IX/X-bp) as expected from the high level of amino acid sequence homology. The functional difference in ligand recognition from IX/X-bp must reside in the amino acid differences. A continuity of different amino acid residues located from the C-terminal of the second alpha-helix to the following loop forms the local conformational difference in this region between the two proteins. This loop participates in the formation of the concave surface between the two subunits, the putative binding site for the Gla-domain (gamma-carboxyglutamic acid-containing domain) of the coagulation factors. Another difference between the two proteins is in the relative disposition of subunits A and B. When the B subunits are superimposed, about a 6 degrees rotation is required for the superposition of the A subunits. A calcium ion links the second alpha-helix region to the C-terminal tail in each subunit and helps to stabilize the structure for Gla-domain binding. The interface created by the central loop swapping in the dimer IX-bp is almost identical with that seen within the monomeric C-type CRDs. This dimer forms as the result of the amino acid deletion in the linker region of the central loop of the original C-type lectins. Such a dimerization disrupts the lectin active site and creates a Gla-domain binding site, imparting functional diversity.
- National Institute of Agrobiological Resources, Tsukuba Science City, Ibaraki, 305-8602, Japan. mizuno@abr.affrc.go.jp
Organizational Affiliation: