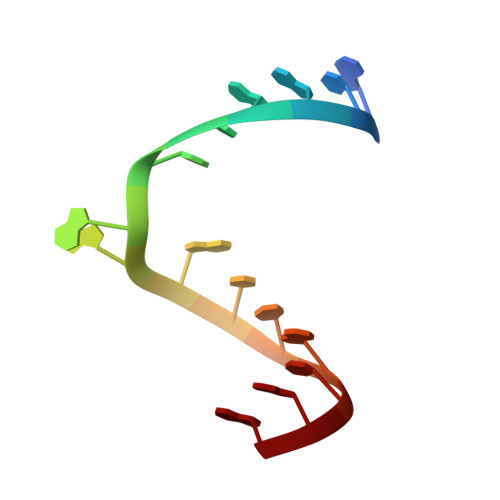
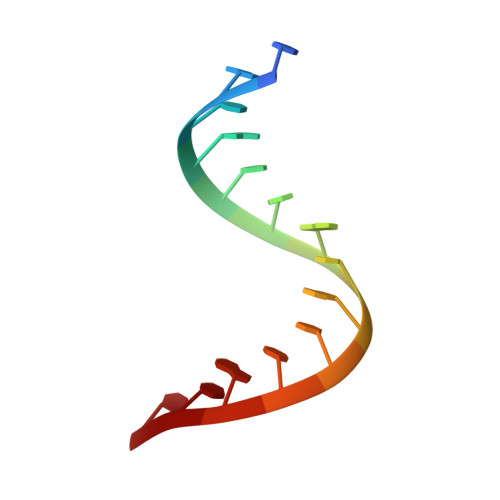
Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Zhao, F., Zhao, Q., Blount, K.F., Han, Q., Tor, Y., Hermann, T.(2005) Angew Chem Int Ed Engl 44: 5329-5334
- PubMed: 16037995
- DOI: https://doi.org/10.1002/anie.200500903
- Primary Citation of Related Structures:
1ZX7, 1ZZ5, 2A04 - Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, 92093, USA.
Organizational Affiliation: