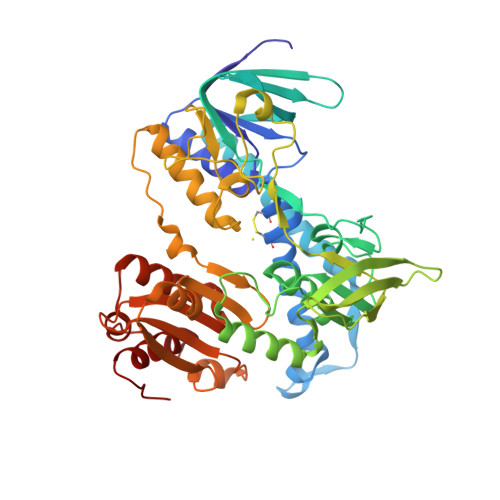
How Dihydrolipoamide Dehydrogenase-binding Protein Binds Dihydrolipoamide Dehydrogenase in the Human Pyruvate Dehydrogenase Complex.
Ciszak, E.M., Makal, A., Hong, Y.S., Vettaikkorumakankauv, A.K., Korotchkina, L.G., Patel, M.S.(2006) J Biological Chem 281: 648-655
- PubMed: 16263718
- DOI: https://doi.org/10.1074/jbc.M507850200
- Primary Citation of Related Structures:
1ZY8 - PubMed Abstract:
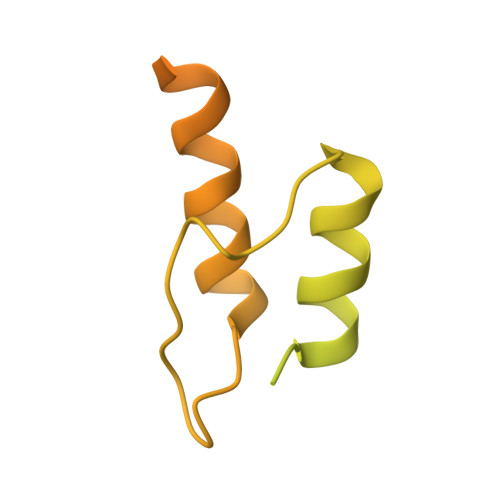
The dihydrolipoamide dehydrogenase-binding protein (E3BP) and the dihydrolipoamide acetyltransferase (E2) component enzyme form the structural core of the human pyruvate dehydrogenase complex by providing the binding sites for two other component proteins, dihydrolipoamide dehydrogenase (E3) and pyruvate dehydrogenase (E1), as well as pyruvate dehydrogenase kinases and phosphatases. Despite a high similarity between the primary structures of E3BP and E2, the E3-binding domain of human E3BP is highly specific to human E3, whereas the E1-binding domain of human E2 is highly specific to human E1. In this study, we characterized binding of human E3 to the E3-binding domain of E3BP by x-ray crystallography at 2.6-angstroms resolution, and we used this structural information to interpret the specificity for selective binding. Two subunits of E3 form a single recognition site for the E3-binding domain of E3BP through their hydrophobic interface. The hydrophobic residues Pro133, Pro154, and Ile157 in the E3-binding domain of E3BP insert themselves into the surface of both E3 polypeptide chains. Numerous ionic and hydrogen bonds between the residues of three interacting polypeptide chains adjacent to the central hydrophobic patch add to the stability of the subcomplex. The specificity of pairing for human E3BP with E3 is interpreted from its subcomplex structure to be most likely due to conformational rigidity of the binding fragment of the E3-binding domain of E3BP and its exquisite amino acid match with the E3 target interface.
- Laboratory for Structural Biology, National Space Science and Technology Center, University of Alabama in Huntsville, 35805, USA. ciszakE@uah.edu
Organizational Affiliation: