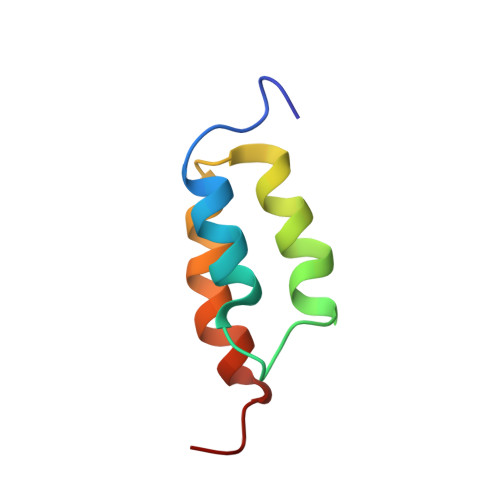
Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
He, Y., Yeh, D.C., Alexander, P., Bryan, P.N., Orban, J.(2005) Biochemistry 44: 14055-14061
- PubMed: 16245921
- DOI: https://doi.org/10.1021/bi051232j
- Primary Citation of Related Structures:
1ZXG, 1ZXH - PubMed Abstract:
We describe here the solution NMR structures of two IgG binding domains with highly homologous sequences but different three-dimensional structures. The proteins, G311 and A219, are derived from the IgG binding domains of their wild-type counterparts, protein G and protein A, respectively. Through a series of site-directed mutations and phage display selections, the sequences of G311 and A219 were designed to converge to a point of high-level sequence identity while keeping their respective wild-type tertiary folds. Structures of both artificially evolved sequences were determined by NMR spectroscopy. The main chain fold of G311 can be superimposed on the wild-type alpha/beta protein G structure with a backbone rmsd of 1.4 A, and the A219 structure can be overlaid on the wild-type three-alpha-helix protein A fold also with a backbone rmsd of 1.4 A. The structure of G311, in particular, accommodates a large number of mutational changes without undergoing a change in the overall fold of the main chain. The structural differences are maintained despite a high level (59%) of sequence identity. These proteins serve as starting points for further experiments that will probe basic concepts of protein folding and conformational switching.
- Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, 9600 Gudelsky Drive, Rockville, Maryland 20850, USA.
Organizational Affiliation: