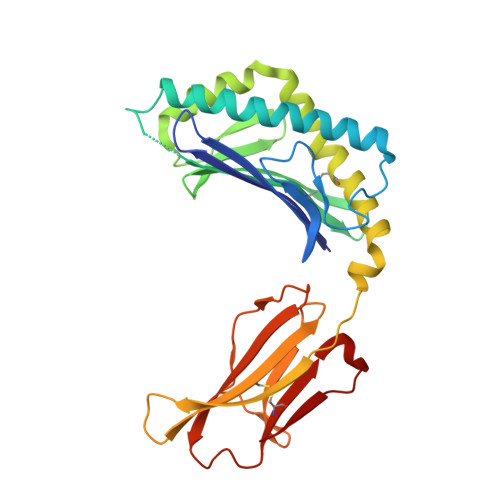
Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation
Giabbai, B., Sidobre, S., Crispin, M.M.D., Sanchez Ruiz, Y., Bachi, A., Kronenberg, M., Wilson, I.A., Degano, M.(2005) J Immunol 175: 977-984
- PubMed: 16002697
- DOI: https://doi.org/10.4049/jimmunol.175.2.977
- Primary Citation of Related Structures:
1ZHN - PubMed Abstract:
NKT cells are immunoregulatory lymphocytes whose activation is triggered by the recognition of lipid Ags in the context of the CD1d molecules by the TCR. In this study we present the crystal structure to 2.8 A of mouse CD1d bound to phosphatidylcholine. The interactions between the ligand acyl chains and the CD1d molecule define the structural and chemical requirements for the binding of lipid Ags to CD1d. The orientation of the polar headgroup toward the C terminus of the alpha1 helix provides a rationale for the structural basis for the observed Valpha chain bias in invariant NKT cells. The contribution of the ligand to the protein surface suggests a likely mode of recognition of lipid Ags by the NKT cell TCR.
- Biocrystallography Unit and Mass Spectrometry Unit, DIBIT San Raffaele Scientific Institute, via Olgettina 58, 20132 Milan, Italy.
Organizational Affiliation: