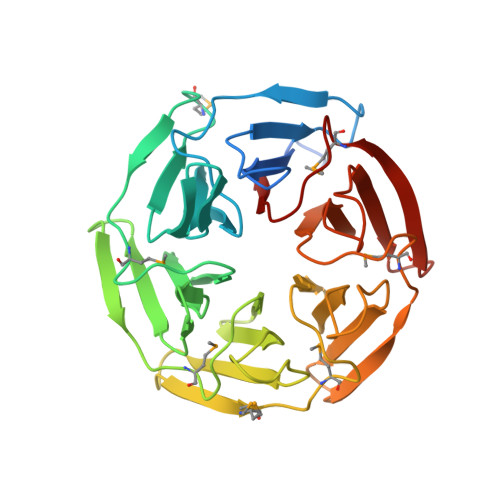
Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Beamer, L.J., Li, X., Bottoms, C.A., Hannink, M.(2005) Acta Crystallogr D Biol Crystallogr 61: 1335-1342
- PubMed: 16204884
- DOI: https://doi.org/10.1107/S0907444905022626
- Primary Citation of Related Structures:
1ZGK - PubMed Abstract:
The Kelch repeat is a common sequence motif in eukaryotic genomes and is approximately 50 amino acids in length. The structure of the Kelch domain of the human Keap1 protein has previously been determined at 1.85 Angstrom, showing that each Kelch repeat forms one blade of a six-bladed beta-propeller. Here, use of 1.35 Angstrom SAD data for de novo structure determination of the Kelch domain and for refinement at atomic resolution is described. The high quality and resolution of the diffraction data and phase information allows a detailed analysis of the role of solvent in the structure of the Kelch repeat. Ten structurally conserved water molecules are identified in each blade of the Kelch beta-propeller. These appear to play distinct structural roles that include lining the central channel of the propeller, interacting with residues in loops between strands of the blade and making contacts with conserved residues in the Kelch repeat. Furthermore, we identify a conserved C-H...pi hydrogen bond between two key residues in the consensus Kelch repeat. This analysis extends our understanding of the structural roles of conserved residues in the Kelch repeat and highlights the potential role of solvent in maintaining the fold of this common eukaryotic structural motif.
- Department of Biochemistry, University of Missouri-Columbia, Columbia, MO 65211, USA. beamerl@missouri.edu
Organizational Affiliation: