Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Hsu, H.C., Stillman, B., Xu, R.M.(2005) Proc Natl Acad Sci U S A 102: 8519-8524
- PubMed: 15937111
- DOI: https://doi.org/10.1073/pnas.0502946102
- Primary Citation of Related Structures:
1ZBX - PubMed Abstract:
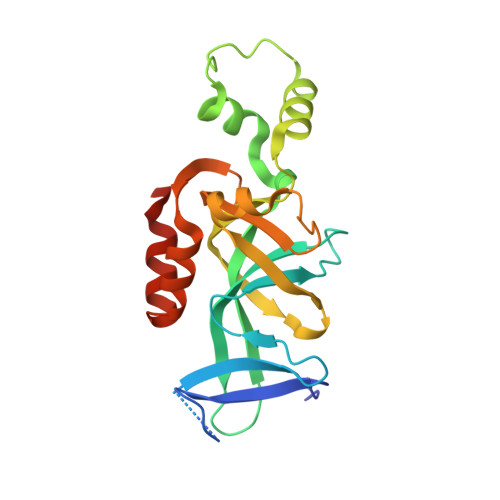
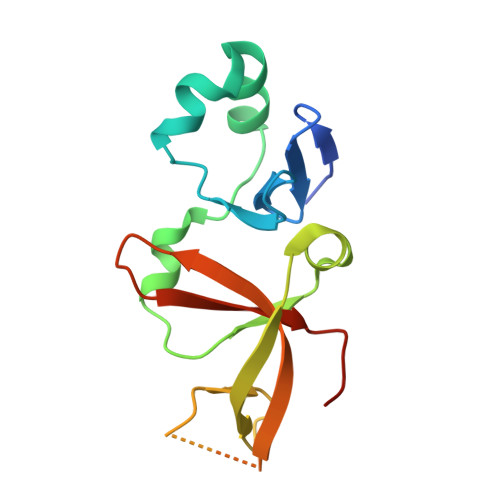
The interaction between silence information regulator 1 protein (Sir1p) and origin recognition complex 1 protein (Orc1p), the largest subunit of the origin recognition complex, plays an important role in the establishment of transcriptional silencing at the cryptic mating-type gene loci in Saccharomyces cerevisiae. Sir1p binds the N-terminal region of Orc1p encompassing a Bromo-adjacent homology (BAH) domain found in various chromatin-associated proteins. To understand the molecular mechanism of Sir protein recruitment, we have determined a 2.5-A cocrystal structure of the N-terminal domain of Orc1p in complex with the Orc1p-interacting domain of Sir1p. The structure reveals that Sir1p Orc1p-interacting domain has a bilobal structure: an alpha/beta N-terminal lobe and a C-terminal lobe resembling the Tudor domain royal family fold. The N-terminal lobe of Sir1p binds in a shallow groove between a helical subdomain and the BAH domain of Orc1p. The structure provides a mechanistic understanding of Orc1p-Sir1p interaction specificity, as well as insights into protein-protein interactions involving BAH domains in general.
- Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.
Organizational Affiliation: