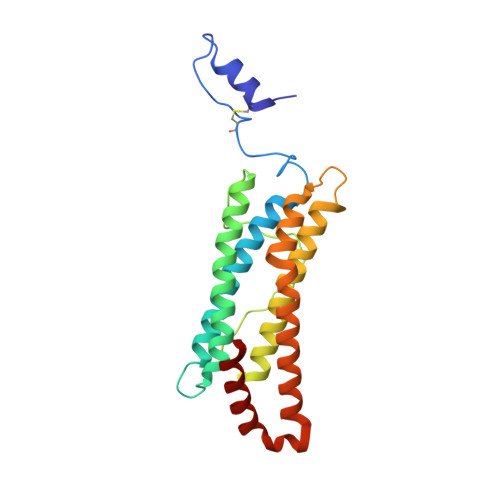
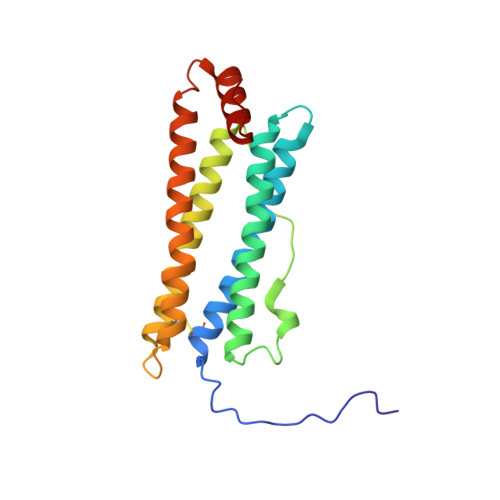
Crystal structure of a secreted insect ferritin reveals a symmetrical arrangement of heavy and light chains.
Hamburger, A.E., West, A.P., Hamburger, Z.A., Hamburger, P., Bjorkman, P.J.(2005) J Mol Biology 349: 558-569
- PubMed: 15896348
- DOI: https://doi.org/10.1016/j.jmb.2005.03.074
- Primary Citation of Related Structures:
1Z6O - PubMed Abstract:
Ferritins are iron storage proteins made of 24 subunits forming a hollow spherical shell. Vertebrate ferritins contain varying ratios of heavy (H) and light (L) chains; however, known ferritin structures include only one type of chain and have octahedral symmetry. Here, we report the 1.9A structure of a secreted insect ferritin from Trichoplusia ni, which reveals equal numbers of H and L chains arranged with tetrahedral symmetry. The H/L-chain interface includes complementary features responsible for ordered assembly of the subunits. The H chain contains a ferroxidase active site resembling that of vertebrate H chains with an endogenous, bound iron atom. The L chain lacks the residues that form a putative iron core nucleation site in vertebrate L chains. Instead, a possible nucleation site is observed at the L chain 3-fold pore. The structure also reveals inter- and intrasubunit disulfide bonds, mostly in the extended N-terminal regions unique to insect ferritins. The symmetrical arrangement of H and L chains and the disulfide crosslinks reflect adaptations of insect ferritin to its role as a secreted protein.
- Division of Biology 114-96, California Institute of Technology, Pasadena, CA 91125, USA.
Organizational Affiliation: